| Full name: phosphoribosylformylglycinamidine synthase | Alias Symbol: PURL|FGARAT|KIAA0361|GATD8 | ||
| Type: protein-coding gene | Cytoband: 17p13.1 | ||
| Entrez ID: 5198 | HGNC ID: HGNC:8863 | Ensembl Gene: ENSG00000178921 | OMIM ID: 602133 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PFAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PFAS | 5198 | 213302_at | 0.5163 | 0.2647 | |
| GSE20347 | PFAS | 5198 | 213302_at | 0.6584 | 0.0041 | |
| GSE23400 | PFAS | 5198 | 213302_at | 0.5467 | 0.0000 | |
| GSE26886 | PFAS | 5198 | 213302_at | 1.3487 | 0.0000 | |
| GSE29001 | PFAS | 5198 | 213302_at | 0.4830 | 0.0834 | |
| GSE38129 | PFAS | 5198 | 213302_at | 0.6684 | 0.0000 | |
| GSE45670 | PFAS | 5198 | 213302_at | 0.6367 | 0.0016 | |
| GSE53622 | PFAS | 5198 | 73080 | 0.7343 | 0.0000 | |
| GSE53624 | PFAS | 5198 | 73080 | 0.6769 | 0.0000 | |
| GSE63941 | PFAS | 5198 | 213302_at | 1.1955 | 0.0108 | |
| GSE77861 | PFAS | 5198 | 213302_at | 0.5362 | 0.0557 | |
| GSE97050 | PFAS | 5198 | A_23_P402610 | 0.7604 | 0.1662 | |
| SRP007169 | PFAS | 5198 | RNAseq | 1.8817 | 0.0001 | |
| SRP008496 | PFAS | 5198 | RNAseq | 1.9733 | 0.0000 | |
| SRP064894 | PFAS | 5198 | RNAseq | 0.5330 | 0.0345 | |
| SRP133303 | PFAS | 5198 | RNAseq | 0.3498 | 0.0194 | |
| SRP159526 | PFAS | 5198 | RNAseq | 0.9467 | 0.0033 | |
| SRP193095 | PFAS | 5198 | RNAseq | 0.5572 | 0.0000 | |
| SRP219564 | PFAS | 5198 | RNAseq | 0.9088 | 0.0550 | |
| TCGA | PFAS | 5198 | RNAseq | 0.0904 | 0.1739 |
Upregulated datasets: 4; Downregulated datasets: 0.
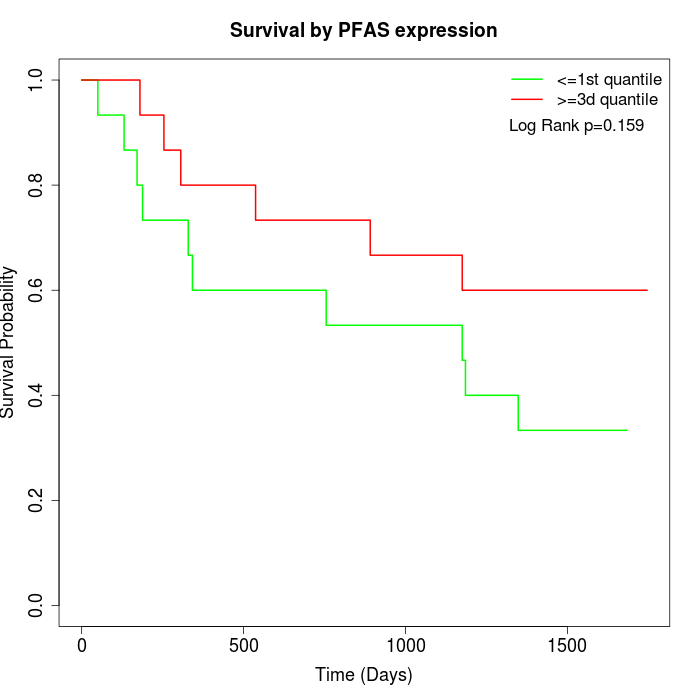
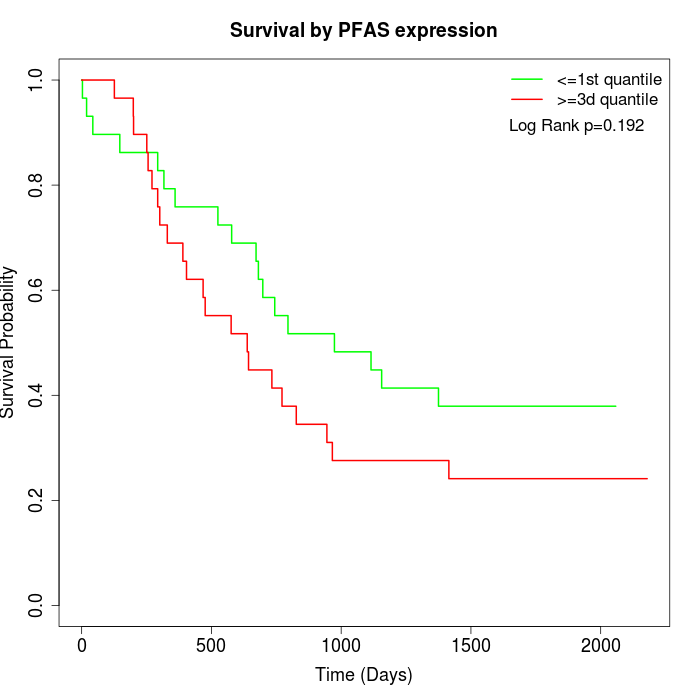
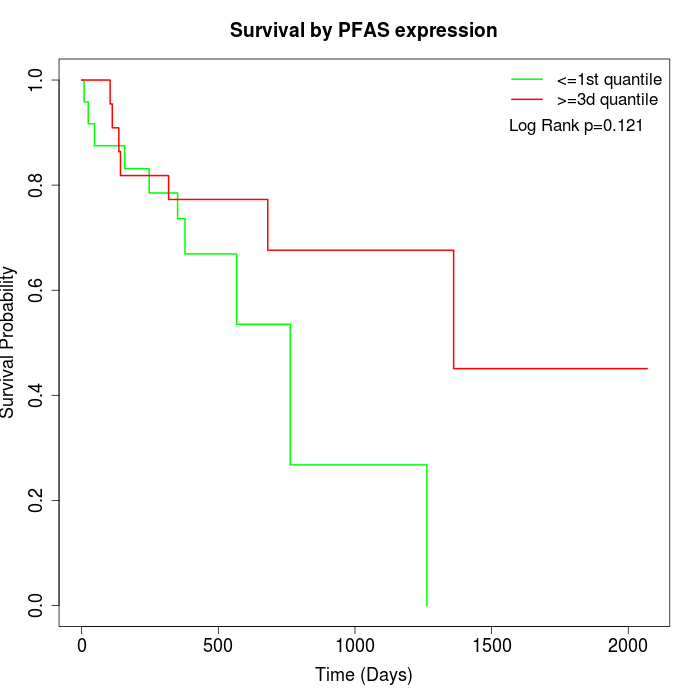
Survival by PFAS expression:
Note: Click image to view full size file.
Copy number change of PFAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PFAS | 5198 | 4 | 2 | 24 | |
| GSE20123 | PFAS | 5198 | 4 | 3 | 23 | |
| GSE43470 | PFAS | 5198 | 1 | 6 | 36 | |
| GSE46452 | PFAS | 5198 | 34 | 1 | 24 | |
| GSE47630 | PFAS | 5198 | 7 | 1 | 32 | |
| GSE54993 | PFAS | 5198 | 4 | 3 | 63 | |
| GSE54994 | PFAS | 5198 | 5 | 8 | 40 | |
| GSE60625 | PFAS | 5198 | 4 | 0 | 7 | |
| GSE74703 | PFAS | 5198 | 1 | 3 | 32 | |
| GSE74704 | PFAS | 5198 | 2 | 1 | 17 | |
| TCGA | PFAS | 5198 | 17 | 21 | 58 |
Total number of gains: 83; Total number of losses: 49; Total Number of normals: 356.
Somatic mutations of PFAS:
Generating mutation plots.
Highly correlated genes for PFAS:
Showing top 20/1758 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PFAS | MDM2 | 0.797949 | 3 | 0 | 3 |
| PFAS | L3MBTL3 | 0.754797 | 3 | 0 | 3 |
| PFAS | PET117 | 0.740784 | 4 | 0 | 3 |
| PFAS | XPO5 | 0.733836 | 6 | 0 | 6 |
| PFAS | ZCCHC3 | 0.732679 | 4 | 0 | 4 |
| PFAS | UCK2 | 0.72932 | 3 | 0 | 3 |
| PFAS | MCM8 | 0.726256 | 7 | 0 | 7 |
| PFAS | ZNF512 | 0.725106 | 3 | 0 | 3 |
| PFAS | XPNPEP3 | 0.716286 | 3 | 0 | 3 |
| PFAS | FMNL2 | 0.71483 | 5 | 0 | 5 |
| PFAS | PPIL1 | 0.71287 | 6 | 0 | 5 |
| PFAS | ZBTB9 | 0.70919 | 6 | 0 | 5 |
| PFAS | EME1 | 0.70869 | 6 | 0 | 5 |
| PFAS | MASTL | 0.70471 | 5 | 0 | 5 |
| PFAS | KIF14 | 0.703792 | 11 | 0 | 11 |
| PFAS | CDCA2 | 0.703652 | 6 | 0 | 5 |
| PFAS | DBF4 | 0.702525 | 10 | 0 | 9 |
| PFAS | RBL1 | 0.701018 | 8 | 0 | 7 |
| PFAS | NEDD1 | 0.700857 | 6 | 0 | 6 |
| PFAS | NUF2 | 0.698567 | 8 | 0 | 7 |
For details and further investigation, click here