| Full name: phosphoglycerate kinase 2 | Alias Symbol: PGKPS|PGK-2 | ||
| Type: protein-coding gene | Cytoband: 6p12.3 | ||
| Entrez ID: 5232 | HGNC ID: HGNC:8898 | Ensembl Gene: ENSG00000170950 | OMIM ID: 172270 |
| Drug and gene relationship at DGIdb | |||
Expression of PGK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGK2 | 5232 | 217009_at | 0.0669 | 0.8338 | |
| GSE20347 | PGK2 | 5232 | 217009_at | -0.0388 | 0.5828 | |
| GSE23400 | PGK2 | 5232 | 217009_at | -0.0047 | 0.8807 | |
| GSE26886 | PGK2 | 5232 | 217009_at | 0.0064 | 0.9625 | |
| GSE29001 | PGK2 | 5232 | 217009_at | -0.1190 | 0.3042 | |
| GSE38129 | PGK2 | 5232 | 217009_at | -0.1031 | 0.1434 | |
| GSE45670 | PGK2 | 5232 | 217009_at | 0.0325 | 0.7554 | |
| GSE53622 | PGK2 | 5232 | 84395 | 0.0428 | 0.8786 | |
| GSE53624 | PGK2 | 5232 | 84395 | -0.0369 | 0.6738 | |
| GSE63941 | PGK2 | 5232 | 217009_at | 0.1520 | 0.2456 | |
| GSE77861 | PGK2 | 5232 | 217009_at | -0.0598 | 0.6600 |
Upregulated datasets: 0; Downregulated datasets: 0.
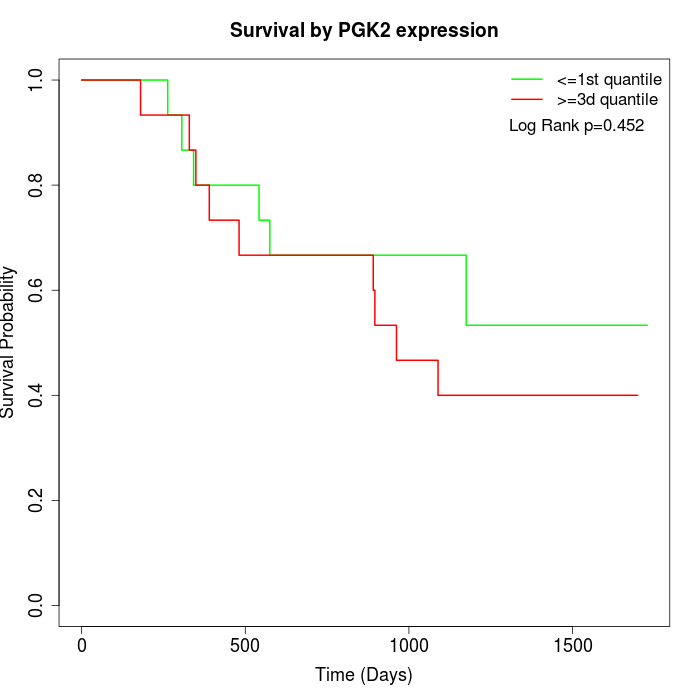
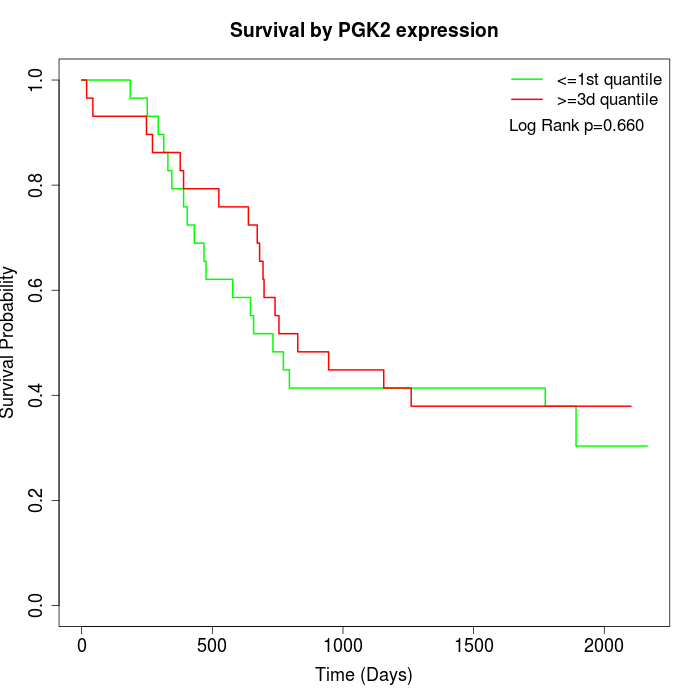
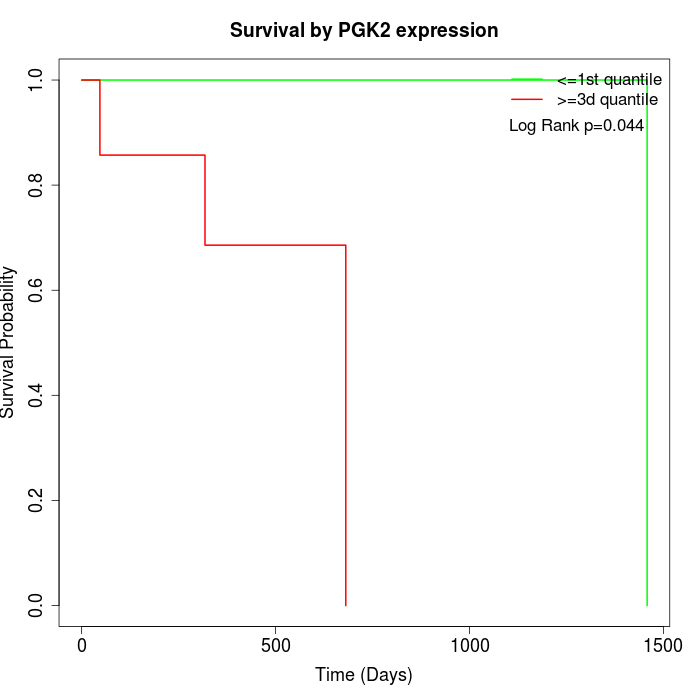
Survival by PGK2 expression:
Note: Click image to view full size file.
Copy number change of PGK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGK2 | 5232 | 5 | 2 | 23 | |
| GSE20123 | PGK2 | 5232 | 5 | 2 | 23 | |
| GSE43470 | PGK2 | 5232 | 5 | 0 | 38 | |
| GSE46452 | PGK2 | 5232 | 2 | 10 | 47 | |
| GSE47630 | PGK2 | 5232 | 7 | 5 | 28 | |
| GSE54993 | PGK2 | 5232 | 3 | 1 | 66 | |
| GSE54994 | PGK2 | 5232 | 8 | 4 | 41 | |
| GSE60625 | PGK2 | 5232 | 0 | 3 | 8 | |
| GSE74703 | PGK2 | 5232 | 5 | 0 | 31 | |
| GSE74704 | PGK2 | 5232 | 2 | 1 | 17 | |
| TCGA | PGK2 | 5232 | 21 | 12 | 63 |
Total number of gains: 63; Total number of losses: 40; Total Number of normals: 385.
Somatic mutations of PGK2:
Generating mutation plots.
Highly correlated genes for PGK2:
Showing top 20/116 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGK2 | IQSEC3 | 0.763821 | 3 | 0 | 3 |
| PGK2 | VGF | 0.726288 | 3 | 0 | 3 |
| PGK2 | IL22 | 0.649084 | 4 | 0 | 3 |
| PGK2 | FOXD3 | 0.627264 | 3 | 0 | 3 |
| PGK2 | PCSK2 | 0.623615 | 3 | 0 | 3 |
| PGK2 | AQP6 | 0.6145 | 4 | 0 | 3 |
| PGK2 | CEBPE | 0.610848 | 5 | 0 | 5 |
| PGK2 | MUC6 | 0.606105 | 4 | 0 | 3 |
| PGK2 | COX6A2 | 0.605293 | 4 | 0 | 3 |
| PGK2 | GUCA1B | 0.599114 | 4 | 0 | 3 |
| PGK2 | LINC00482 | 0.597728 | 3 | 0 | 3 |
| PGK2 | STAB2 | 0.597078 | 3 | 0 | 3 |
| PGK2 | FGF5 | 0.59609 | 3 | 0 | 3 |
| PGK2 | SOX15 | 0.595959 | 3 | 0 | 3 |
| PGK2 | PAEP | 0.59244 | 5 | 0 | 4 |
| PGK2 | PLCB3 | 0.590755 | 5 | 0 | 4 |
| PGK2 | OR2J2 | 0.589363 | 4 | 0 | 4 |
| PGK2 | EXTL1 | 0.588676 | 4 | 0 | 3 |
| PGK2 | EHD1 | 0.587737 | 3 | 0 | 3 |
| PGK2 | NEUROD2 | 0.585407 | 3 | 0 | 3 |
For details and further investigation, click here