| Full name: piwi like RNA-mediated gene silencing 4 | Alias Symbol: FLJ36156|HIWI2|Miwi2 | ||
| Type: protein-coding gene | Cytoband: 11q21 | ||
| Entrez ID: 143689 | HGNC ID: HGNC:18444 | Ensembl Gene: ENSG00000134627 | OMIM ID: 610315 |
| Drug and gene relationship at DGIdb | |||
Expression of PIWIL4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIWIL4 | 143689 | 230480_at | -0.3102 | 0.5494 | |
| GSE26886 | PIWIL4 | 143689 | 230480_at | 0.6635 | 0.0063 | |
| GSE45670 | PIWIL4 | 143689 | 230480_at | -0.3660 | 0.1426 | |
| GSE53622 | PIWIL4 | 143689 | 14975 | 0.0940 | 0.2814 | |
| GSE53624 | PIWIL4 | 143689 | 14975 | 0.1753 | 0.0066 | |
| GSE63941 | PIWIL4 | 143689 | 230480_at | -0.8659 | 0.0338 | |
| GSE77861 | PIWIL4 | 143689 | 230480_at | -0.0395 | 0.7031 | |
| GSE97050 | PIWIL4 | 143689 | A_23_P427760 | 0.1571 | 0.4148 | |
| SRP064894 | PIWIL4 | 143689 | RNAseq | -0.0586 | 0.8375 | |
| SRP133303 | PIWIL4 | 143689 | RNAseq | -0.4935 | 0.0682 | |
| SRP159526 | PIWIL4 | 143689 | RNAseq | -0.0680 | 0.8934 | |
| SRP219564 | PIWIL4 | 143689 | RNAseq | -0.1846 | 0.6523 | |
| TCGA | PIWIL4 | 143689 | RNAseq | -1.0942 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
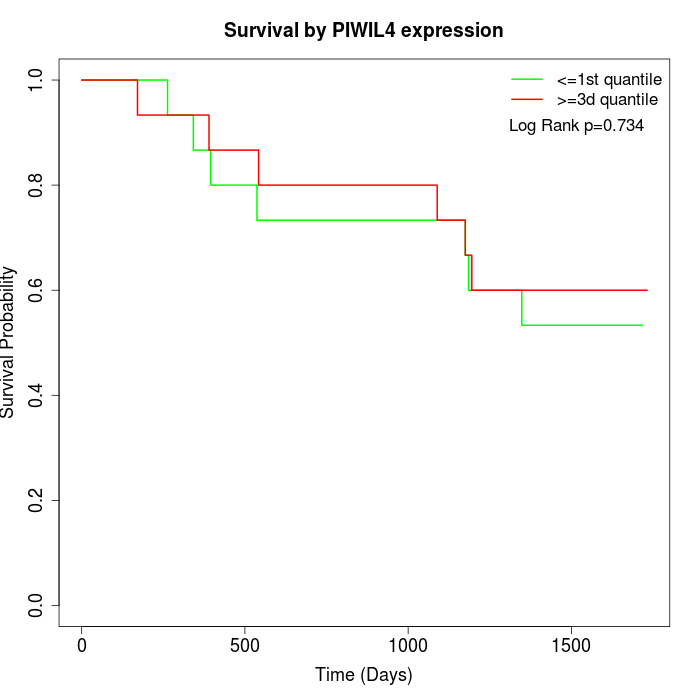
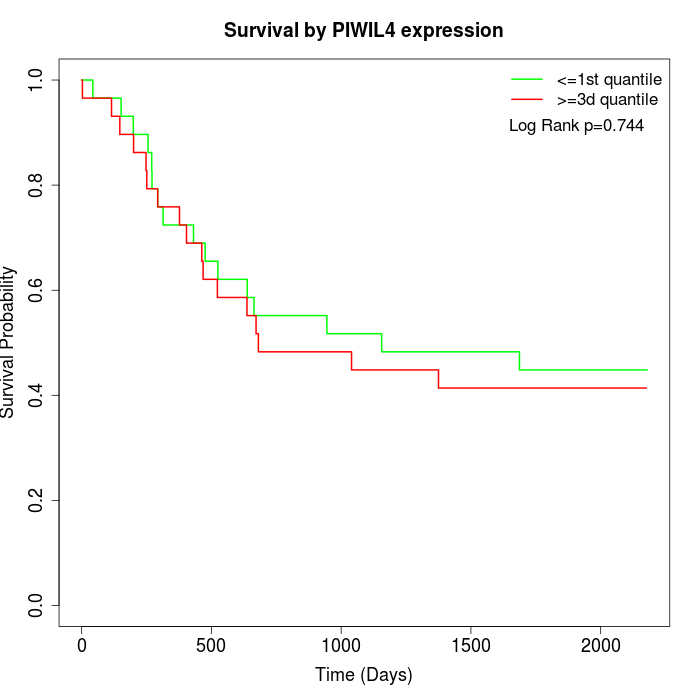
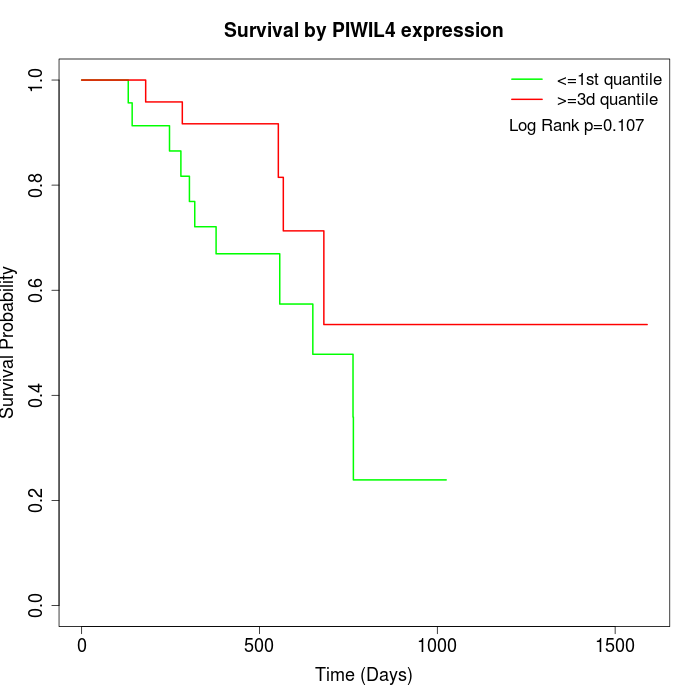
Survival by PIWIL4 expression:
Note: Click image to view full size file.
Copy number change of PIWIL4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIWIL4 | 143689 | 3 | 9 | 18 | |
| GSE20123 | PIWIL4 | 143689 | 3 | 9 | 18 | |
| GSE43470 | PIWIL4 | 143689 | 2 | 5 | 36 | |
| GSE46452 | PIWIL4 | 143689 | 5 | 24 | 30 | |
| GSE47630 | PIWIL4 | 143689 | 2 | 19 | 19 | |
| GSE54993 | PIWIL4 | 143689 | 8 | 2 | 60 | |
| GSE54994 | PIWIL4 | 143689 | 7 | 18 | 28 | |
| GSE60625 | PIWIL4 | 143689 | 0 | 3 | 8 | |
| GSE74703 | PIWIL4 | 143689 | 1 | 4 | 31 | |
| GSE74704 | PIWIL4 | 143689 | 3 | 5 | 12 | |
| TCGA | PIWIL4 | 143689 | 11 | 43 | 42 |
Total number of gains: 45; Total number of losses: 141; Total Number of normals: 302.
Somatic mutations of PIWIL4:
Generating mutation plots.
Highly correlated genes for PIWIL4:
Showing top 20/48 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIWIL4 | TBX5-AS1 | 0.702705 | 3 | 0 | 3 |
| PIWIL4 | CPQ | 0.669539 | 3 | 0 | 3 |
| PIWIL4 | ZNF500 | 0.652181 | 3 | 0 | 3 |
| PIWIL4 | BEND6 | 0.635887 | 4 | 0 | 4 |
| PIWIL4 | HOMER3 | 0.627421 | 3 | 0 | 3 |
| PIWIL4 | IKBKB | 0.624216 | 3 | 0 | 3 |
| PIWIL4 | LYRM9 | 0.622292 | 3 | 0 | 3 |
| PIWIL4 | TBX3 | 0.612894 | 3 | 0 | 3 |
| PIWIL4 | IVD | 0.605452 | 3 | 0 | 3 |
| PIWIL4 | NAT14 | 0.603194 | 3 | 0 | 3 |
| PIWIL4 | TBX2 | 0.601242 | 3 | 0 | 3 |
| PIWIL4 | ENTPD1 | 0.599966 | 4 | 0 | 4 |
| PIWIL4 | PLCL2 | 0.59529 | 5 | 0 | 4 |
| PIWIL4 | FAM110B | 0.594855 | 4 | 0 | 4 |
| PIWIL4 | NTNG2 | 0.594129 | 3 | 0 | 3 |
| PIWIL4 | PHF21A | 0.592588 | 3 | 0 | 3 |
| PIWIL4 | FOXL1 | 0.590936 | 3 | 0 | 3 |
| PIWIL4 | SHC2 | 0.590625 | 4 | 0 | 3 |
| PIWIL4 | EID2B | 0.589499 | 5 | 0 | 3 |
| PIWIL4 | PAX3 | 0.588931 | 3 | 0 | 3 |
For details and further investigation, click here