| Full name: phospholipid scramblase 1 | Alias Symbol: MMTRA1B | ||
| Type: protein-coding gene | Cytoband: 3q24 | ||
| Entrez ID: 5359 | HGNC ID: HGNC:9092 | Ensembl Gene: ENSG00000188313 | OMIM ID: 604170 |
| Drug and gene relationship at DGIdb | |||
Expression of PLSCR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLSCR1 | 5359 | 202446_s_at | 0.2429 | 0.4079 | |
| GSE20347 | PLSCR1 | 5359 | 202446_s_at | 0.4247 | 0.1352 | |
| GSE23400 | PLSCR1 | 5359 | 202446_s_at | 0.4330 | 0.0004 | |
| GSE26886 | PLSCR1 | 5359 | 202446_s_at | -1.0036 | 0.0011 | |
| GSE29001 | PLSCR1 | 5359 | 202446_s_at | 0.7121 | 0.0287 | |
| GSE38129 | PLSCR1 | 5359 | 202446_s_at | 0.4001 | 0.0600 | |
| GSE45670 | PLSCR1 | 5359 | 202446_s_at | 0.8180 | 0.0006 | |
| GSE53622 | PLSCR1 | 5359 | 28768 | 0.3439 | 0.0010 | |
| GSE53624 | PLSCR1 | 5359 | 28768 | 0.4044 | 0.0001 | |
| GSE63941 | PLSCR1 | 5359 | 202446_s_at | 0.6172 | 0.6657 | |
| GSE77861 | PLSCR1 | 5359 | 202446_s_at | 0.1227 | 0.7450 | |
| GSE97050 | PLSCR1 | 5359 | A_23_P69109 | 0.2854 | 0.6268 | |
| SRP007169 | PLSCR1 | 5359 | RNAseq | 0.0578 | 0.8950 | |
| SRP008496 | PLSCR1 | 5359 | RNAseq | -0.1256 | 0.7059 | |
| SRP064894 | PLSCR1 | 5359 | RNAseq | 0.6451 | 0.0179 | |
| SRP133303 | PLSCR1 | 5359 | RNAseq | 0.4881 | 0.0373 | |
| SRP159526 | PLSCR1 | 5359 | RNAseq | -0.3397 | 0.5160 | |
| SRP193095 | PLSCR1 | 5359 | RNAseq | 0.4011 | 0.0271 | |
| SRP219564 | PLSCR1 | 5359 | RNAseq | 0.4973 | 0.4421 | |
| TCGA | PLSCR1 | 5359 | RNAseq | 0.2618 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 1.
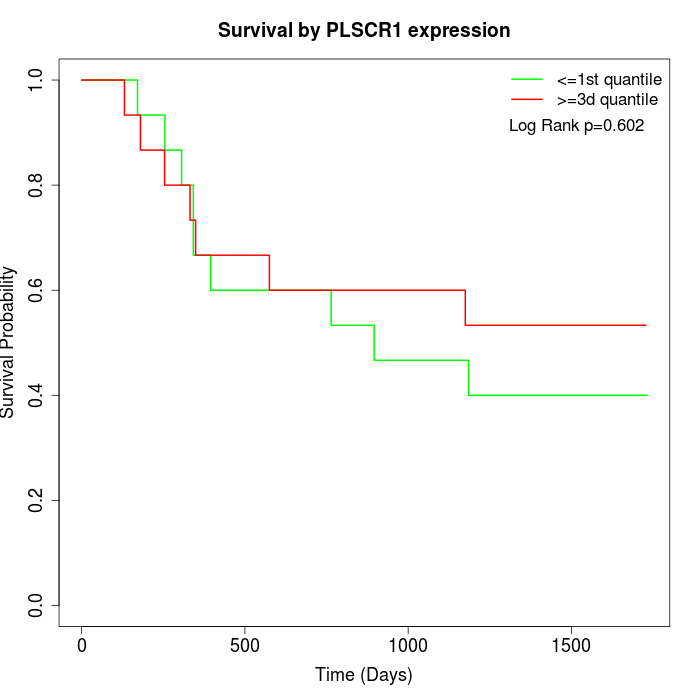
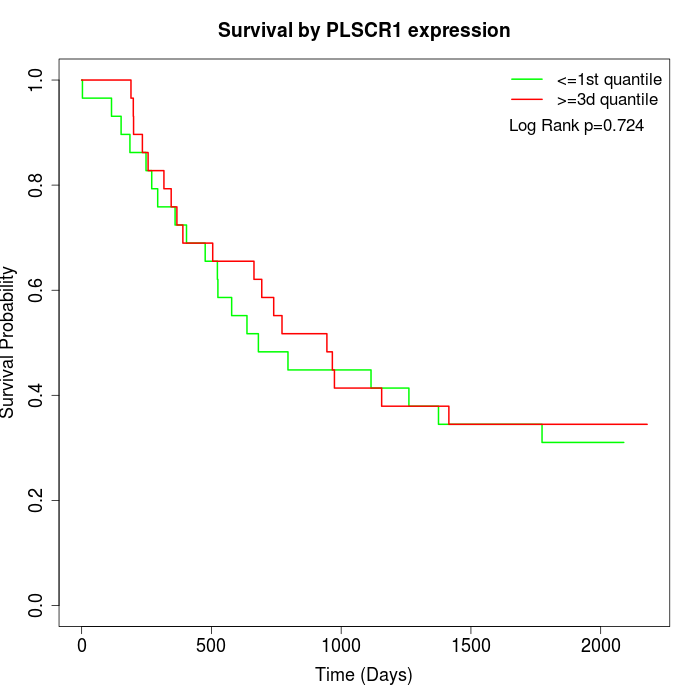
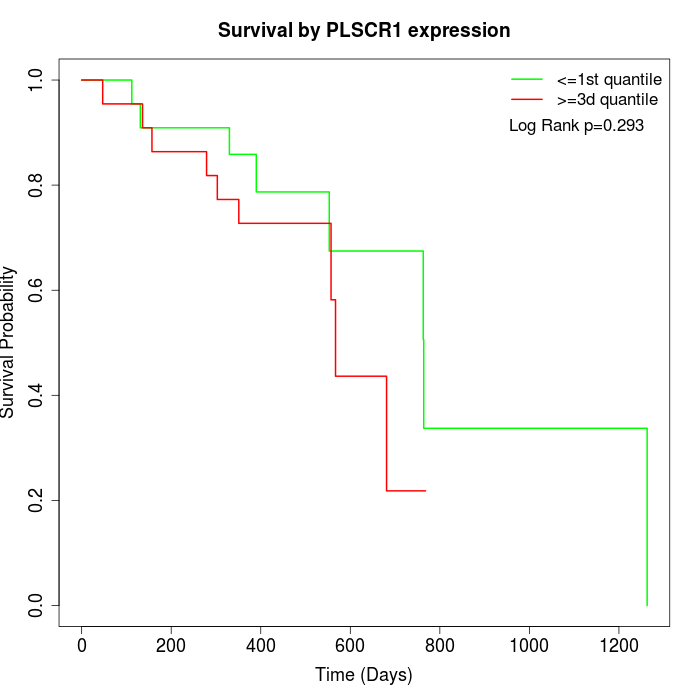
Survival by PLSCR1 expression:
Note: Click image to view full size file.
Copy number change of PLSCR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLSCR1 | 5359 | 22 | 0 | 8 | |
| GSE20123 | PLSCR1 | 5359 | 22 | 0 | 8 | |
| GSE43470 | PLSCR1 | 5359 | 23 | 0 | 20 | |
| GSE46452 | PLSCR1 | 5359 | 16 | 3 | 40 | |
| GSE47630 | PLSCR1 | 5359 | 19 | 3 | 18 | |
| GSE54993 | PLSCR1 | 5359 | 1 | 9 | 60 | |
| GSE54994 | PLSCR1 | 5359 | 34 | 1 | 18 | |
| GSE60625 | PLSCR1 | 5359 | 0 | 6 | 5 | |
| GSE74703 | PLSCR1 | 5359 | 20 | 0 | 16 | |
| GSE74704 | PLSCR1 | 5359 | 15 | 0 | 5 | |
| TCGA | PLSCR1 | 5359 | 67 | 2 | 27 |
Total number of gains: 239; Total number of losses: 24; Total Number of normals: 225.
Somatic mutations of PLSCR1:
Generating mutation plots.
Highly correlated genes for PLSCR1:
Showing top 20/285 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLSCR1 | RNF213 | 0.824157 | 3 | 0 | 3 |
| PLSCR1 | CCDC85C | 0.82303 | 3 | 0 | 3 |
| PLSCR1 | PARP9 | 0.741436 | 8 | 0 | 8 |
| PLSCR1 | ELP4 | 0.73758 | 3 | 0 | 3 |
| PLSCR1 | DGCR2 | 0.728193 | 3 | 0 | 3 |
| PLSCR1 | QSOX2 | 0.726912 | 3 | 0 | 3 |
| PLSCR1 | CDCA7 | 0.71841 | 4 | 0 | 3 |
| PLSCR1 | CD80 | 0.717484 | 3 | 0 | 3 |
| PLSCR1 | SMAD1 | 0.716752 | 3 | 0 | 3 |
| PLSCR1 | SYMPK | 0.708981 | 3 | 0 | 3 |
| PLSCR1 | RAB27A | 0.704608 | 3 | 0 | 3 |
| PLSCR1 | RBM8A | 0.703916 | 3 | 0 | 3 |
| PLSCR1 | BTG1 | 0.703622 | 3 | 0 | 3 |
| PLSCR1 | DDX60L | 0.698622 | 4 | 0 | 3 |
| PLSCR1 | PLEKHA7 | 0.694712 | 3 | 0 | 3 |
| PLSCR1 | SMAP2 | 0.691849 | 3 | 0 | 3 |
| PLSCR1 | RNF2 | 0.691061 | 3 | 0 | 3 |
| PLSCR1 | YTHDF3 | 0.689907 | 3 | 0 | 3 |
| PLSCR1 | TRIM47 | 0.689318 | 3 | 0 | 3 |
| PLSCR1 | AKAP17A | 0.686992 | 3 | 0 | 3 |
For details and further investigation, click here