| Full name: peroxisome proliferator activated receptor delta | Alias Symbol: NUC1|NUCII|FAAR|NR1C2 | ||
| Type: protein-coding gene | Cytoband: 6p21.31 | ||
| Entrez ID: 5467 | HGNC ID: HGNC:9235 | Ensembl Gene: ENSG00000112033 | OMIM ID: 600409 |
| Drug and gene relationship at DGIdb | |||
PPARD involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway | |
| hsa04310 | Wnt signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05221 | Acute myeloid leukemia |
Expression of PPARD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPARD | 5467 | 37152_at | 0.2648 | 0.6468 | |
| GSE20347 | PPARD | 5467 | 37152_at | -0.2794 | 0.0950 | |
| GSE23400 | PPARD | 5467 | 37152_at | 0.0071 | 0.9363 | |
| GSE26886 | PPARD | 5467 | 37152_at | 0.2481 | 0.5330 | |
| GSE29001 | PPARD | 5467 | 37152_at | -0.2298 | 0.6431 | |
| GSE38129 | PPARD | 5467 | 37152_at | -0.2111 | 0.2575 | |
| GSE45670 | PPARD | 5467 | 37152_at | 0.3723 | 0.0657 | |
| GSE53622 | PPARD | 5467 | 81459 | -0.1227 | 0.3271 | |
| GSE53624 | PPARD | 5467 | 81459 | -0.3094 | 0.0004 | |
| GSE63941 | PPARD | 5467 | 37152_at | -0.3438 | 0.3693 | |
| GSE77861 | PPARD | 5467 | 37152_at | -0.2044 | 0.4388 | |
| GSE97050 | PPARD | 5467 | A_23_P214681 | 0.4314 | 0.2930 | |
| SRP007169 | PPARD | 5467 | RNAseq | -2.0796 | 0.0000 | |
| SRP008496 | PPARD | 5467 | RNAseq | -1.8819 | 0.0000 | |
| SRP064894 | PPARD | 5467 | RNAseq | -0.1473 | 0.5361 | |
| SRP133303 | PPARD | 5467 | RNAseq | -0.0309 | 0.8967 | |
| SRP159526 | PPARD | 5467 | RNAseq | -0.6042 | 0.0306 | |
| SRP193095 | PPARD | 5467 | RNAseq | -0.1413 | 0.4665 | |
| SRP219564 | PPARD | 5467 | RNAseq | -0.4389 | 0.3533 | |
| TCGA | PPARD | 5467 | RNAseq | 0.0236 | 0.7321 |
Upregulated datasets: 0; Downregulated datasets: 2.
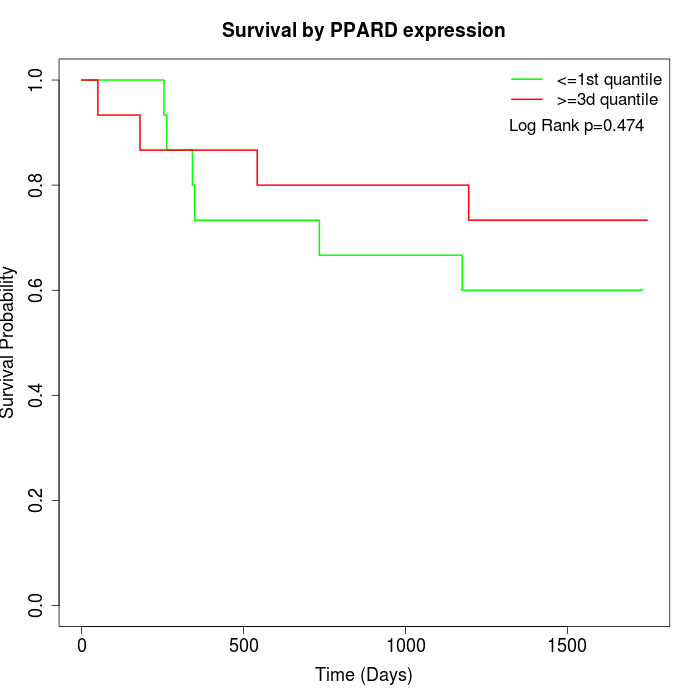
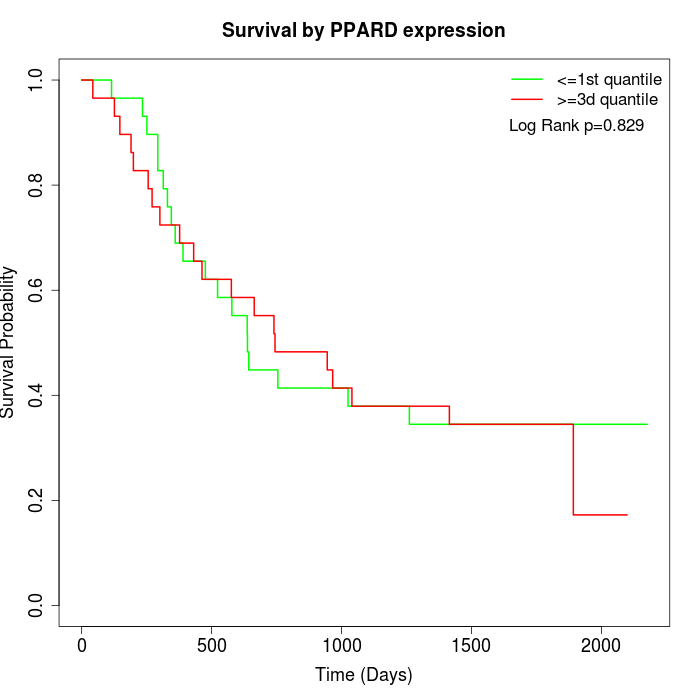
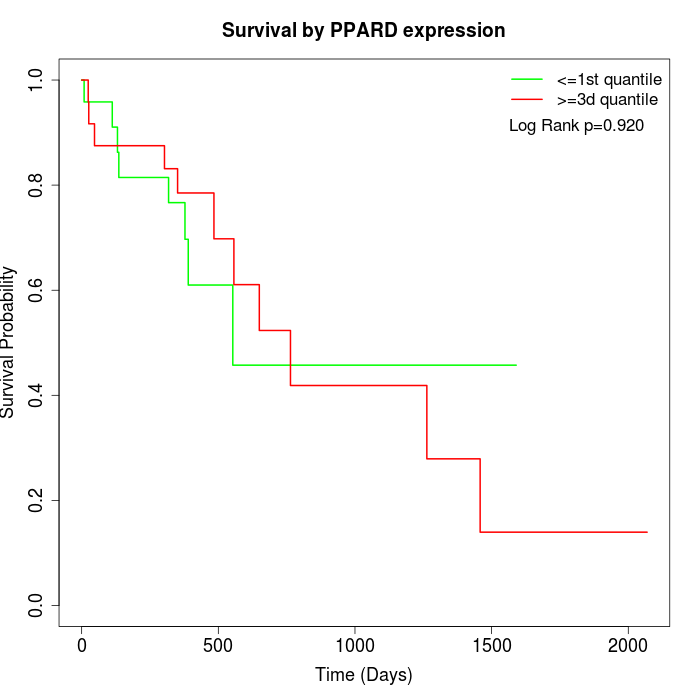
Survival by PPARD expression:
Note: Click image to view full size file.
Copy number change of PPARD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPARD | 5467 | 5 | 0 | 25 | |
| GSE20123 | PPARD | 5467 | 5 | 0 | 25 | |
| GSE43470 | PPARD | 5467 | 5 | 1 | 37 | |
| GSE46452 | PPARD | 5467 | 2 | 9 | 48 | |
| GSE47630 | PPARD | 5467 | 8 | 4 | 28 | |
| GSE54993 | PPARD | 5467 | 3 | 1 | 66 | |
| GSE54994 | PPARD | 5467 | 11 | 4 | 38 | |
| GSE60625 | PPARD | 5467 | 1 | 0 | 10 | |
| GSE74703 | PPARD | 5467 | 5 | 0 | 31 | |
| GSE74704 | PPARD | 5467 | 2 | 0 | 18 | |
| TCGA | PPARD | 5467 | 16 | 15 | 65 |
Total number of gains: 63; Total number of losses: 34; Total Number of normals: 391.
Somatic mutations of PPARD:
Generating mutation plots.
Highly correlated genes for PPARD:
Showing top 20/380 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPARD | SLC35F5 | 0.713159 | 3 | 0 | 3 |
| PPARD | TTC39B | 0.688304 | 3 | 0 | 3 |
| PPARD | FXYD5 | 0.683906 | 3 | 0 | 3 |
| PPARD | PDE12 | 0.68215 | 3 | 0 | 3 |
| PPARD | CMIP | 0.679218 | 4 | 0 | 4 |
| PPARD | ALKBH3 | 0.671038 | 3 | 0 | 3 |
| PPARD | SNCG | 0.666925 | 3 | 0 | 3 |
| PPARD | GPR45 | 0.666027 | 3 | 0 | 3 |
| PPARD | RMDN1 | 0.664954 | 3 | 0 | 3 |
| PPARD | SFN | 0.659056 | 11 | 0 | 10 |
| PPARD | KPTN | 0.653788 | 4 | 0 | 4 |
| PPARD | LY6K | 0.653254 | 4 | 0 | 4 |
| PPARD | BPNT1 | 0.65156 | 3 | 0 | 3 |
| PPARD | UNC5B | 0.651028 | 5 | 0 | 3 |
| PPARD | PRSS53 | 0.650362 | 3 | 0 | 3 |
| PPARD | CXCL16 | 0.647421 | 3 | 0 | 3 |
| PPARD | VSNL1 | 0.645826 | 4 | 0 | 3 |
| PPARD | ANKRD1 | 0.64316 | 3 | 0 | 3 |
| PPARD | LACTB | 0.643021 | 4 | 0 | 3 |
| PPARD | WNT5A | 0.638479 | 4 | 0 | 3 |
For details and further investigation, click here