| Full name: pregnancy specific beta-1-glycoprotein 6 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19q13.31 | ||
| Entrez ID: 5675 | HGNC ID: HGNC:9523 | Ensembl Gene: ENSG00000170848 | OMIM ID: 176395 |
| Drug and gene relationship at DGIdb | |||
Expression of PSG6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PSG6 | 5675 | 239910_at | -0.0762 | 0.7045 | |
| GSE20347 | PSG6 | 5675 | 208106_x_at | -0.2185 | 0.0521 | |
| GSE23400 | PSG6 | 5675 | 208106_x_at | -0.3368 | 0.0000 | |
| GSE26886 | PSG6 | 5675 | 208106_x_at | -0.5275 | 0.0001 | |
| GSE29001 | PSG6 | 5675 | 208106_x_at | -0.1608 | 0.4676 | |
| GSE38129 | PSG6 | 5675 | 208106_x_at | -0.2782 | 0.0042 | |
| GSE45670 | PSG6 | 5675 | 208106_x_at | 0.0440 | 0.8000 | |
| GSE63941 | PSG6 | 5675 | 208106_x_at | 0.0858 | 0.9016 | |
| GSE77861 | PSG6 | 5675 | 208106_x_at | -0.0470 | 0.7005 | |
| GSE97050 | PSG6 | A_21_P0000191 | -0.4709 | 0.5008 | ||
| TCGA | PSG6 | 5675 | RNAseq | 2.0461 | 0.3318 |
Upregulated datasets: 0; Downregulated datasets: 0.
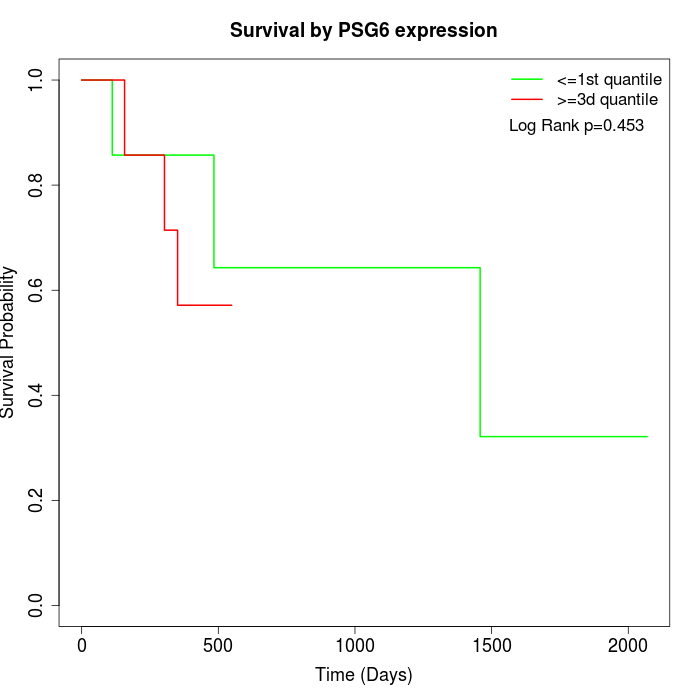
Survival by PSG6 expression:
Note: Click image to view full size file.
Copy number change of PSG6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PSG6 | 5675 | 6 | 5 | 19 | |
| GSE20123 | PSG6 | 5675 | 5 | 4 | 21 | |
| GSE43470 | PSG6 | 5675 | 5 | 11 | 27 | |
| GSE46452 | PSG6 | 5675 | 46 | 1 | 12 | |
| GSE47630 | PSG6 | 5675 | 8 | 6 | 26 | |
| GSE54993 | PSG6 | 5675 | 17 | 4 | 49 | |
| GSE54994 | PSG6 | 5675 | 7 | 12 | 34 | |
| GSE60625 | PSG6 | 5675 | 9 | 0 | 2 | |
| GSE74703 | PSG6 | 5675 | 5 | 7 | 24 | |
| GSE74704 | PSG6 | 5675 | 4 | 2 | 14 | |
| TCGA | PSG6 | 5675 | 14 | 15 | 67 |
Total number of gains: 126; Total number of losses: 67; Total Number of normals: 295.
Somatic mutations of PSG6:
Generating mutation plots.
Highly correlated genes for PSG6:
Showing top 20/811 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PSG6 | MUC5AC | 0.716516 | 4 | 0 | 4 |
| PSG6 | GRM6 | 0.713926 | 6 | 0 | 6 |
| PSG6 | WWP2 | 0.704999 | 3 | 0 | 3 |
| PSG6 | GCG | 0.702944 | 3 | 0 | 3 |
| PSG6 | PSG2 | 0.700375 | 7 | 0 | 6 |
| PSG6 | PRL | 0.688085 | 4 | 0 | 4 |
| PSG6 | LY6G6C | 0.687697 | 4 | 0 | 3 |
| PSG6 | CATSPER3 | 0.68694 | 3 | 0 | 3 |
| PSG6 | PSG1 | 0.68671 | 7 | 0 | 6 |
| PSG6 | HRC | 0.684953 | 6 | 0 | 5 |
| PSG6 | RNF17 | 0.684627 | 5 | 0 | 4 |
| PSG6 | IGLON5 | 0.682351 | 3 | 0 | 3 |
| PSG6 | OTOR | 0.681129 | 4 | 0 | 4 |
| PSG6 | CETN1 | 0.680018 | 4 | 0 | 4 |
| PSG6 | IL17RC | 0.679384 | 7 | 0 | 6 |
| PSG6 | SPATA19 | 0.67932 | 3 | 0 | 3 |
| PSG6 | NCR1 | 0.678757 | 6 | 0 | 5 |
| PSG6 | FCN2 | 0.675953 | 5 | 0 | 4 |
| PSG6 | SLC26A3 | 0.673628 | 3 | 0 | 3 |
| PSG6 | SPACA1 | 0.671889 | 4 | 0 | 4 |
For details and further investigation, click here