| Full name: protein tyrosine phosphatase non-receptor type 2 | Alias Symbol: TCELLPTP|TC-PTP|TCPTP | ||
| Type: protein-coding gene | Cytoband: 18p11.21 | ||
| Entrez ID: 5771 | HGNC ID: HGNC:9650 | Ensembl Gene: ENSG00000175354 | OMIM ID: 176887 |
| Drug and gene relationship at DGIdb | |||
PTPN2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04630 | Jak-STAT signaling pathway |
Expression of PTPN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PTPN2 | 5771 | 213136_at | 0.6132 | 0.1921 | |
| GSE20347 | PTPN2 | 5771 | 213136_at | 0.5571 | 0.0000 | |
| GSE23400 | PTPN2 | 5771 | 213136_at | 0.5438 | 0.0000 | |
| GSE26886 | PTPN2 | 5771 | 213136_at | 1.4547 | 0.0000 | |
| GSE29001 | PTPN2 | 5771 | 213136_at | 0.6464 | 0.0290 | |
| GSE38129 | PTPN2 | 5771 | 213136_at | 0.7457 | 0.0000 | |
| GSE45670 | PTPN2 | 5771 | 213136_at | 0.3950 | 0.0069 | |
| GSE53622 | PTPN2 | 5771 | 49050 | 0.7138 | 0.0000 | |
| GSE53624 | PTPN2 | 5771 | 49050 | 1.0180 | 0.0000 | |
| GSE63941 | PTPN2 | 5771 | 213136_at | 1.0404 | 0.0335 | |
| GSE77861 | PTPN2 | 5771 | 213136_at | 0.6074 | 0.0057 | |
| GSE97050 | PTPN2 | 5771 | A_32_P1445 | 0.5861 | 0.2307 | |
| SRP007169 | PTPN2 | 5771 | RNAseq | 0.6895 | 0.0496 | |
| SRP008496 | PTPN2 | 5771 | RNAseq | 0.4921 | 0.0275 | |
| SRP064894 | PTPN2 | 5771 | RNAseq | 0.6570 | 0.0003 | |
| SRP133303 | PTPN2 | 5771 | RNAseq | 0.5340 | 0.0012 | |
| SRP159526 | PTPN2 | 5771 | RNAseq | 0.6909 | 0.0004 | |
| SRP193095 | PTPN2 | 5771 | RNAseq | 0.4418 | 0.0000 | |
| SRP219564 | PTPN2 | 5771 | RNAseq | 0.4430 | 0.0785 | |
| TCGA | PTPN2 | 5771 | RNAseq | 0.2429 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
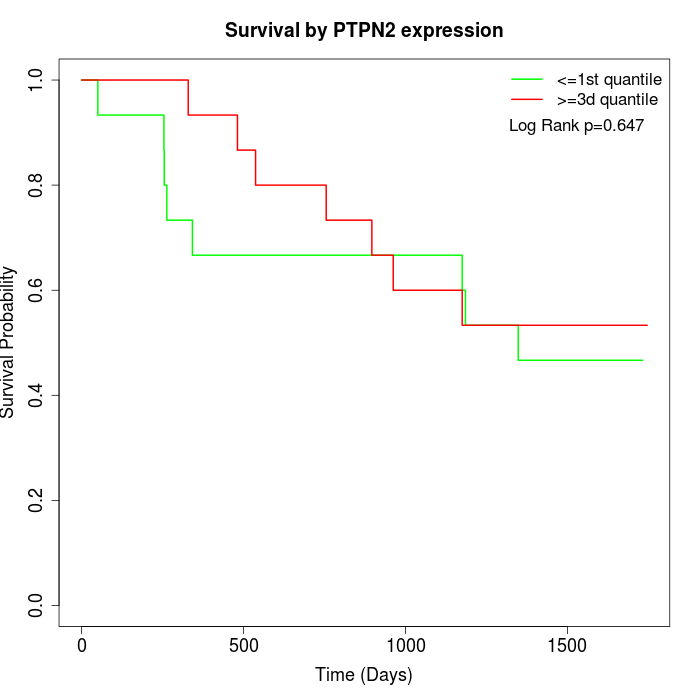
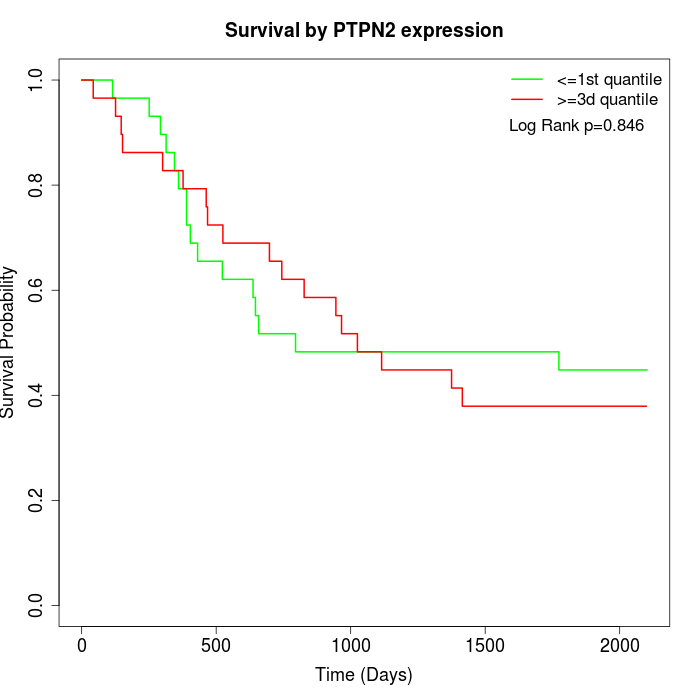
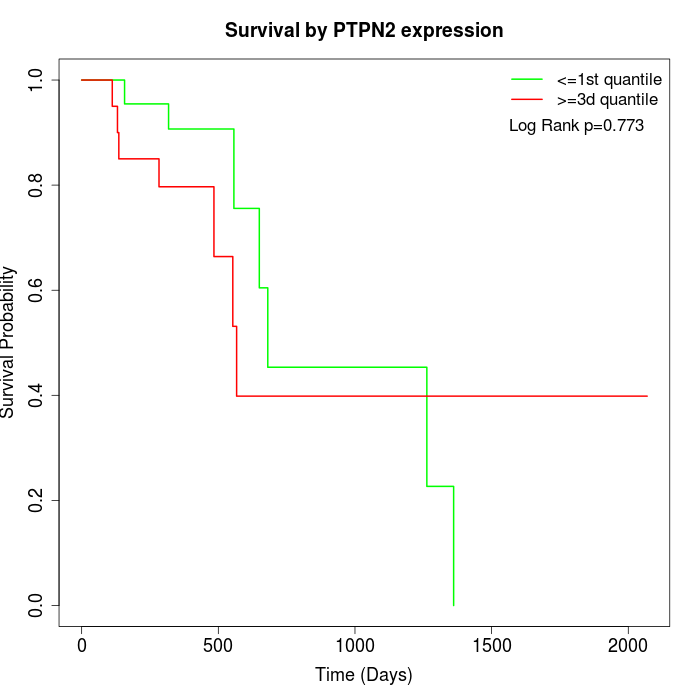
Survival by PTPN2 expression:
Note: Click image to view full size file.
Copy number change of PTPN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PTPN2 | 5771 | 5 | 3 | 22 | |
| GSE20123 | PTPN2 | 5771 | 5 | 3 | 22 | |
| GSE43470 | PTPN2 | 5771 | 0 | 6 | 37 | |
| GSE46452 | PTPN2 | 5771 | 3 | 21 | 35 | |
| GSE47630 | PTPN2 | 5771 | 7 | 17 | 16 | |
| GSE54993 | PTPN2 | 5771 | 6 | 2 | 62 | |
| GSE54994 | PTPN2 | 5771 | 8 | 8 | 37 | |
| GSE60625 | PTPN2 | 5771 | 0 | 4 | 7 | |
| GSE74703 | PTPN2 | 5771 | 0 | 4 | 32 | |
| GSE74704 | PTPN2 | 5771 | 3 | 2 | 15 | |
| TCGA | PTPN2 | 5771 | 29 | 23 | 44 |
Total number of gains: 66; Total number of losses: 93; Total Number of normals: 329.
Somatic mutations of PTPN2:
Generating mutation plots.
Highly correlated genes for PTPN2:
Showing top 20/1784 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PTPN2 | ZSCAN21 | 0.778127 | 3 | 0 | 3 |
| PTPN2 | PHIP | 0.774767 | 5 | 0 | 5 |
| PTPN2 | A4GALT | 0.765215 | 3 | 0 | 3 |
| PTPN2 | ALKBH6 | 0.759257 | 5 | 0 | 5 |
| PTPN2 | LRP4 | 0.758724 | 3 | 0 | 3 |
| PTPN2 | PATL1 | 0.758631 | 3 | 0 | 3 |
| PTPN2 | EXOC6 | 0.755556 | 3 | 0 | 3 |
| PTPN2 | NPFFR1 | 0.738765 | 3 | 0 | 3 |
| PTPN2 | LRRC8C | 0.72796 | 3 | 0 | 3 |
| PTPN2 | FMNL2 | 0.727804 | 7 | 0 | 7 |
| PTPN2 | PGM2L1 | 0.72402 | 4 | 0 | 4 |
| PTPN2 | NOTCH3 | 0.722008 | 3 | 0 | 3 |
| PTPN2 | CSNK2A2 | 0.717582 | 3 | 0 | 3 |
| PTPN2 | RPF2 | 0.717143 | 5 | 0 | 5 |
| PTPN2 | MRPL47 | 0.71192 | 7 | 0 | 6 |
| PTPN2 | CMTM4 | 0.709833 | 4 | 0 | 4 |
| PTPN2 | ZNF121 | 0.706597 | 4 | 0 | 3 |
| PTPN2 | CEP192 | 0.706341 | 12 | 0 | 11 |
| PTPN2 | KRTCAP2 | 0.70561 | 5 | 0 | 5 |
| PTPN2 | INTS4 | 0.704809 | 4 | 0 | 3 |
For details and further investigation, click here