| Full name: casein kinase 2 alpha 2 | Alias Symbol: CSNK2A1|CK2alpha' | ||
| Type: protein-coding gene | Cytoband: 16q21 | ||
| Entrez ID: 1459 | HGNC ID: HGNC:2459 | Ensembl Gene: ENSG00000070770 | OMIM ID: 115442 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CSNK2A2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04310 | Wnt signaling pathway | |
| hsa04520 | Adherens junction | |
| hsa04530 | Tight junction | |
| hsa05168 | Herpes simplex infection |
Expression of CSNK2A2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSNK2A2 | 1459 | 224922_at | 0.4680 | 0.2997 | |
| GSE20347 | CSNK2A2 | 1459 | 203575_at | 0.2899 | 0.0556 | |
| GSE23400 | CSNK2A2 | 1459 | 203575_at | 0.0813 | 0.2167 | |
| GSE26886 | CSNK2A2 | 1459 | 203575_at | 0.6449 | 0.0033 | |
| GSE29001 | CSNK2A2 | 1459 | 203575_at | -0.0818 | 0.7714 | |
| GSE38129 | CSNK2A2 | 1459 | 203575_at | 0.2396 | 0.0757 | |
| GSE45670 | CSNK2A2 | 1459 | 224922_at | 0.2041 | 0.1617 | |
| GSE53622 | CSNK2A2 | 1459 | 101792 | 0.1607 | 0.0599 | |
| GSE53624 | CSNK2A2 | 1459 | 101792 | 0.3664 | 0.0000 | |
| GSE63941 | CSNK2A2 | 1459 | 224922_at | 0.1417 | 0.7073 | |
| GSE77861 | CSNK2A2 | 1459 | 224922_at | 0.3012 | 0.2247 | |
| GSE97050 | CSNK2A2 | 1459 | A_23_P14915 | -0.1022 | 0.6509 | |
| SRP007169 | CSNK2A2 | 1459 | RNAseq | 0.4433 | 0.2164 | |
| SRP008496 | CSNK2A2 | 1459 | RNAseq | 0.3298 | 0.2392 | |
| SRP064894 | CSNK2A2 | 1459 | RNAseq | 0.1942 | 0.2144 | |
| SRP133303 | CSNK2A2 | 1459 | RNAseq | 0.5348 | 0.0000 | |
| SRP159526 | CSNK2A2 | 1459 | RNAseq | 0.3904 | 0.0453 | |
| SRP193095 | CSNK2A2 | 1459 | RNAseq | 0.3126 | 0.0015 | |
| SRP219564 | CSNK2A2 | 1459 | RNAseq | -0.1734 | 0.6249 | |
| TCGA | CSNK2A2 | 1459 | RNAseq | -0.0182 | 0.7169 |
Upregulated datasets: 0; Downregulated datasets: 0.
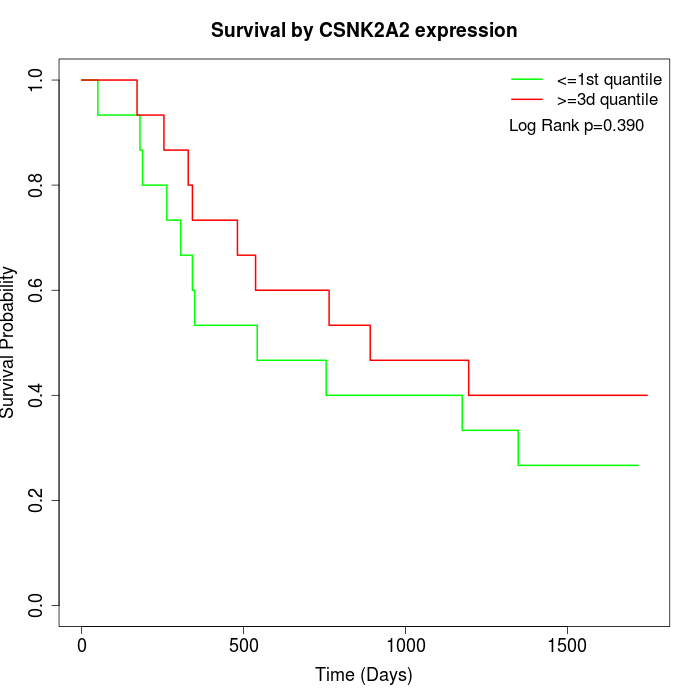
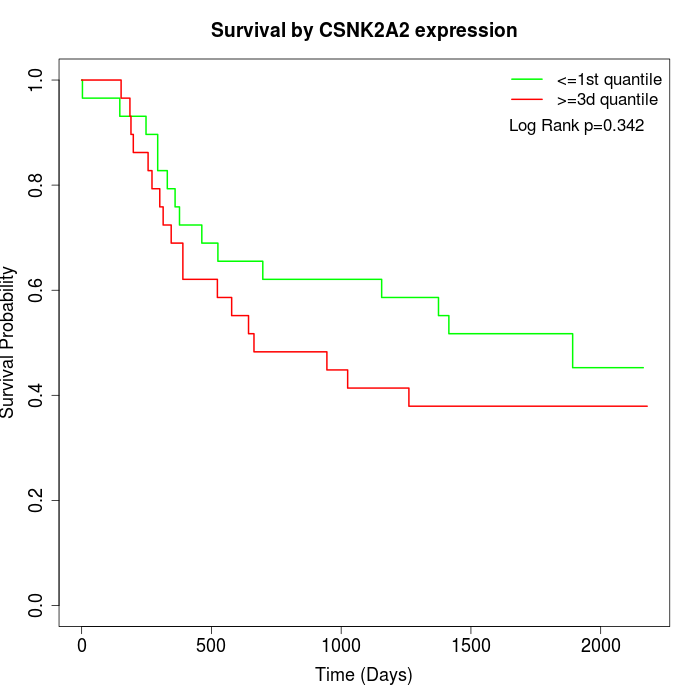
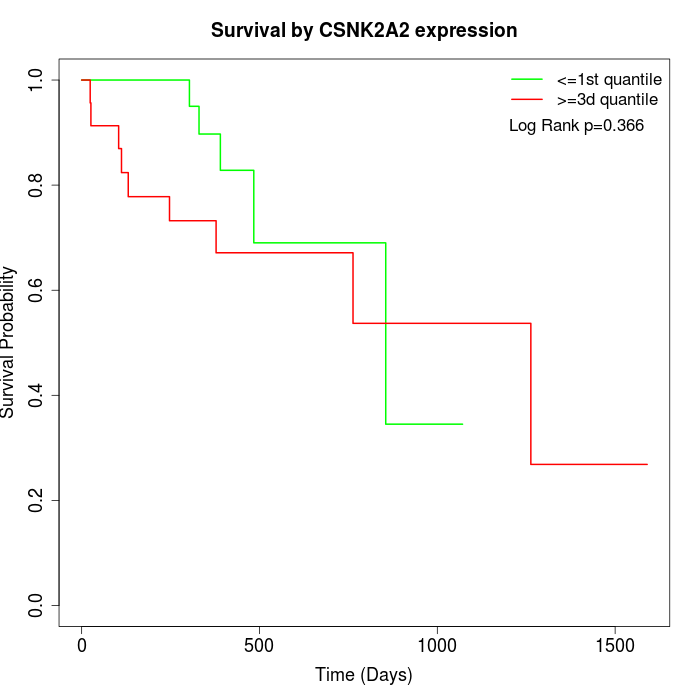
Survival by CSNK2A2 expression:
Note: Click image to view full size file.
Copy number change of CSNK2A2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSNK2A2 | 1459 | 7 | 0 | 23 | |
| GSE20123 | CSNK2A2 | 1459 | 7 | 1 | 22 | |
| GSE43470 | CSNK2A2 | 1459 | 2 | 7 | 34 | |
| GSE46452 | CSNK2A2 | 1459 | 38 | 1 | 20 | |
| GSE47630 | CSNK2A2 | 1459 | 10 | 8 | 22 | |
| GSE54993 | CSNK2A2 | 1459 | 2 | 4 | 64 | |
| GSE54994 | CSNK2A2 | 1459 | 6 | 10 | 37 | |
| GSE60625 | CSNK2A2 | 1459 | 4 | 0 | 7 | |
| GSE74703 | CSNK2A2 | 1459 | 2 | 5 | 29 | |
| GSE74704 | CSNK2A2 | 1459 | 5 | 0 | 15 | |
| TCGA | CSNK2A2 | 1459 | 26 | 11 | 59 |
Total number of gains: 109; Total number of losses: 47; Total Number of normals: 332.
Somatic mutations of CSNK2A2:
Generating mutation plots.
Highly correlated genes for CSNK2A2:
Showing top 20/920 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSNK2A2 | MGAT4B | 0.823825 | 3 | 0 | 3 |
| CSNK2A2 | IRF2BPL | 0.802323 | 3 | 0 | 3 |
| CSNK2A2 | UHRF1BP1 | 0.786085 | 4 | 0 | 4 |
| CSNK2A2 | PPTC7 | 0.776474 | 3 | 0 | 3 |
| CSNK2A2 | ARID2 | 0.771677 | 3 | 0 | 3 |
| CSNK2A2 | C12orf65 | 0.767781 | 3 | 0 | 3 |
| CSNK2A2 | BAG4 | 0.760997 | 3 | 0 | 3 |
| CSNK2A2 | XRCC6 | 0.75953 | 4 | 0 | 3 |
| CSNK2A2 | SEC16A | 0.749367 | 3 | 0 | 3 |
| CSNK2A2 | PITX2 | 0.745127 | 3 | 0 | 3 |
| CSNK2A2 | PXMP4 | 0.744982 | 3 | 0 | 3 |
| CSNK2A2 | LANCL2 | 0.742557 | 4 | 0 | 4 |
| CSNK2A2 | APEH | 0.742529 | 3 | 0 | 3 |
| CSNK2A2 | GLI3 | 0.735248 | 4 | 0 | 4 |
| CSNK2A2 | CCT6A | 0.73092 | 4 | 0 | 3 |
| CSNK2A2 | C12orf4 | 0.72751 | 3 | 0 | 3 |
| CSNK2A2 | PTPN4 | 0.726944 | 3 | 0 | 3 |
| CSNK2A2 | KIF2A | 0.725589 | 3 | 0 | 3 |
| CSNK2A2 | CDC123 | 0.724499 | 4 | 0 | 3 |
| CSNK2A2 | SPNS1 | 0.722027 | 3 | 0 | 3 |
For details and further investigation, click here