| Full name: glycogen phosphorylase B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 20p11.21 | ||
| Entrez ID: 5834 | HGNC ID: HGNC:9723 | Ensembl Gene: ENSG00000100994 | OMIM ID: 138550 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PYGB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04910 | Insulin signaling pathway | |
| hsa04922 | Glucagon signaling pathway | |
| hsa04931 | Insulin resistance |
Expression of PYGB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYGB | 5834 | 201481_s_at | -0.2466 | 0.7589 | |
| GSE20347 | PYGB | 5834 | 201481_s_at | 0.2782 | 0.1717 | |
| GSE23400 | PYGB | 5834 | 201481_s_at | -0.0637 | 0.5482 | |
| GSE26886 | PYGB | 5834 | 201481_s_at | -0.0708 | 0.6955 | |
| GSE29001 | PYGB | 5834 | 201481_s_at | 0.0733 | 0.8344 | |
| GSE38129 | PYGB | 5834 | 201481_s_at | 0.0127 | 0.9672 | |
| GSE45670 | PYGB | 5834 | 201481_s_at | -0.2076 | 0.2206 | |
| GSE53622 | PYGB | 5834 | 101265 | -0.2983 | 0.0154 | |
| GSE53624 | PYGB | 5834 | 101265 | -0.2308 | 0.0071 | |
| GSE63941 | PYGB | 5834 | 201481_s_at | 0.5644 | 0.2383 | |
| GSE77861 | PYGB | 5834 | 201481_s_at | 0.5887 | 0.0224 | |
| GSE97050 | PYGB | 5834 | A_23_P331670 | -0.7122 | 0.2151 | |
| SRP007169 | PYGB | 5834 | RNAseq | -0.3974 | 0.3714 | |
| SRP008496 | PYGB | 5834 | RNAseq | -0.3517 | 0.1663 | |
| SRP064894 | PYGB | 5834 | RNAseq | -0.4297 | 0.0376 | |
| SRP133303 | PYGB | 5834 | RNAseq | -0.0365 | 0.8965 | |
| SRP159526 | PYGB | 5834 | RNAseq | 0.3493 | 0.4240 | |
| SRP193095 | PYGB | 5834 | RNAseq | 0.2890 | 0.1100 | |
| SRP219564 | PYGB | 5834 | RNAseq | -0.0196 | 0.9786 | |
| TCGA | PYGB | 5834 | RNAseq | -0.0971 | 0.2165 |
Upregulated datasets: 0; Downregulated datasets: 0.
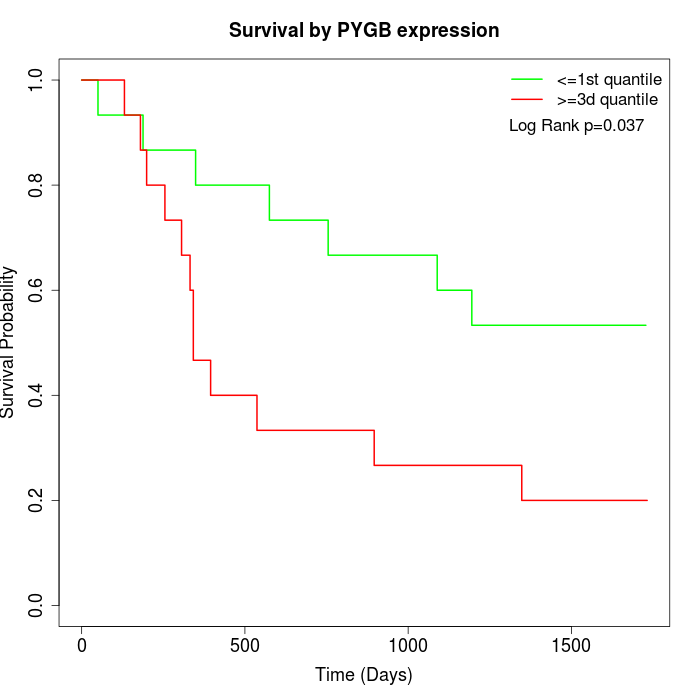
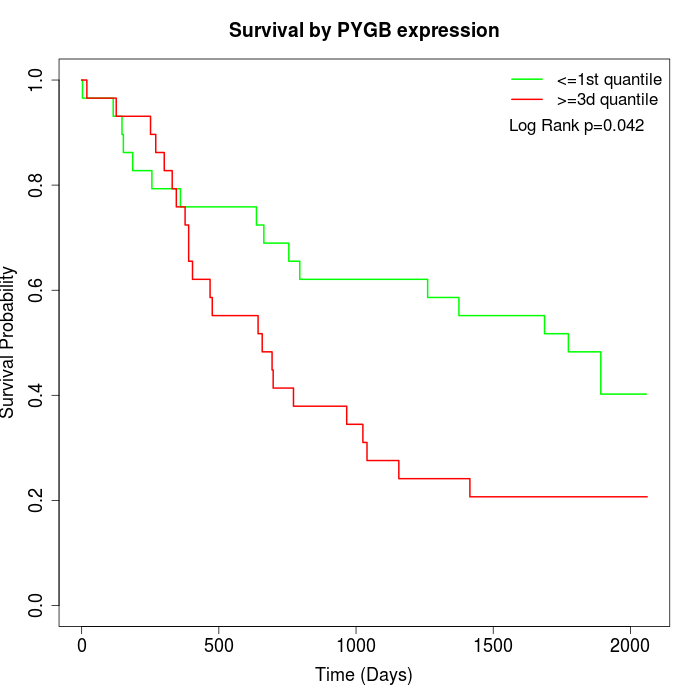
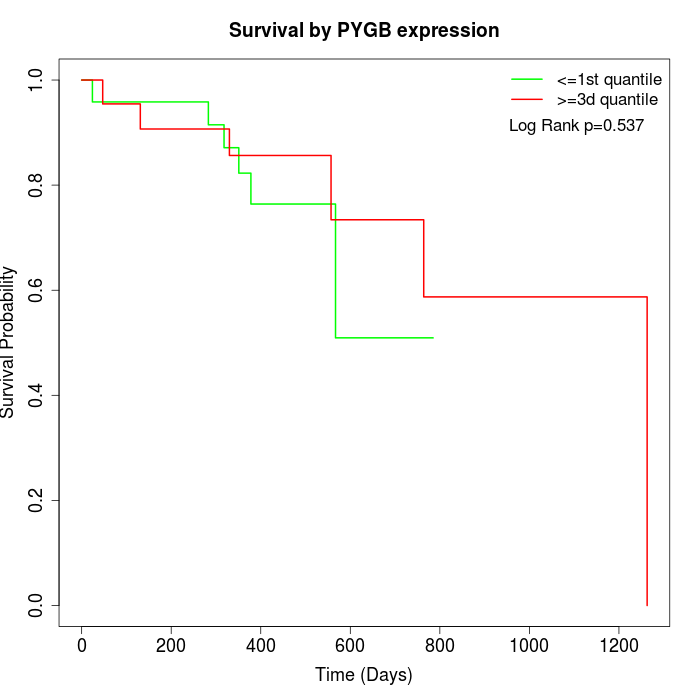
Survival by PYGB expression:
Note: Click image to view full size file.
Copy number change of PYGB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYGB | 5834 | 12 | 2 | 16 | |
| GSE20123 | PYGB | 5834 | 12 | 2 | 16 | |
| GSE43470 | PYGB | 5834 | 11 | 1 | 31 | |
| GSE46452 | PYGB | 5834 | 27 | 1 | 31 | |
| GSE47630 | PYGB | 5834 | 17 | 4 | 19 | |
| GSE54993 | PYGB | 5834 | 0 | 17 | 53 | |
| GSE54994 | PYGB | 5834 | 27 | 1 | 25 | |
| GSE60625 | PYGB | 5834 | 0 | 0 | 11 | |
| GSE74703 | PYGB | 5834 | 10 | 1 | 25 | |
| GSE74704 | PYGB | 5834 | 7 | 1 | 12 | |
| TCGA | PYGB | 5834 | 44 | 10 | 42 |
Total number of gains: 167; Total number of losses: 40; Total Number of normals: 281.
Somatic mutations of PYGB:
Generating mutation plots.
Highly correlated genes for PYGB:
Showing top 20/187 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYGB | PRRT2 | 0.696889 | 3 | 0 | 3 |
| PYGB | ITGB1BP2 | 0.689205 | 4 | 0 | 4 |
| PYGB | PDZD4 | 0.686762 | 3 | 0 | 3 |
| PYGB | AAR2 | 0.678868 | 3 | 0 | 3 |
| PYGB | PHLPP2 | 0.661263 | 3 | 0 | 3 |
| PYGB | CACNA1H | 0.656502 | 5 | 0 | 4 |
| PYGB | CTNNBL1 | 0.655477 | 3 | 0 | 3 |
| PYGB | ENTPD6 | 0.651695 | 7 | 0 | 7 |
| PYGB | FOXF2 | 0.650815 | 7 | 0 | 6 |
| PYGB | COL4A6 | 0.645835 | 6 | 0 | 4 |
| PYGB | ZNF532 | 0.644215 | 3 | 0 | 3 |
| PYGB | UQCC1 | 0.642584 | 3 | 0 | 3 |
| PYGB | MCAM | 0.637423 | 4 | 0 | 4 |
| PYGB | TUBA1A | 0.637019 | 3 | 0 | 3 |
| PYGB | SLC12A2 | 0.634788 | 5 | 0 | 5 |
| PYGB | FNDC5 | 0.634426 | 3 | 0 | 3 |
| PYGB | AP1S2 | 0.630836 | 5 | 0 | 5 |
| PYGB | OLIG2 | 0.627901 | 3 | 0 | 3 |
| PYGB | IQUB | 0.625201 | 3 | 0 | 3 |
| PYGB | CASQ1 | 0.624425 | 5 | 0 | 4 |
For details and further investigation, click here