| Full name: pyridine nucleotide-disulphide oxidoreductase domain 1 | Alias Symbol: DKFZp762G094|FLJ22028 | ||
| Type: protein-coding gene | Cytoband: 12p12.1 | ||
| Entrez ID: 79912 | HGNC ID: HGNC:26162 | Ensembl Gene: ENSG00000121350 | OMIM ID: 617220 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PYROXD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYROXD1 | 79912 | 213878_at | -0.9040 | 0.0323 | |
| GSE20347 | PYROXD1 | 79912 | 219802_at | -0.2414 | 0.3483 | |
| GSE23400 | PYROXD1 | 79912 | 219802_at | -0.1185 | 0.1321 | |
| GSE26886 | PYROXD1 | 79912 | 213878_at | -0.2276 | 0.5576 | |
| GSE29001 | PYROXD1 | 79912 | 219802_at | -0.3123 | 0.1580 | |
| GSE38129 | PYROXD1 | 79912 | 219802_at | -0.4017 | 0.0761 | |
| GSE45670 | PYROXD1 | 79912 | 213878_at | -0.4296 | 0.0030 | |
| GSE53622 | PYROXD1 | 79912 | 2430 | -0.6992 | 0.0000 | |
| GSE53624 | PYROXD1 | 79912 | 2430 | -0.5386 | 0.0000 | |
| GSE63941 | PYROXD1 | 79912 | 213878_at | -0.9055 | 0.2142 | |
| GSE77861 | PYROXD1 | 79912 | 213878_at | -0.3650 | 0.3645 | |
| GSE97050 | PYROXD1 | 79912 | A_33_P3278265 | 0.2148 | 0.3274 | |
| SRP007169 | PYROXD1 | 79912 | RNAseq | -1.6694 | 0.0002 | |
| SRP008496 | PYROXD1 | 79912 | RNAseq | -1.2630 | 0.0001 | |
| SRP064894 | PYROXD1 | 79912 | RNAseq | -0.4583 | 0.0003 | |
| SRP133303 | PYROXD1 | 79912 | RNAseq | -0.1954 | 0.3314 | |
| SRP159526 | PYROXD1 | 79912 | RNAseq | -0.7965 | 0.0003 | |
| SRP193095 | PYROXD1 | 79912 | RNAseq | -0.8053 | 0.0000 | |
| SRP219564 | PYROXD1 | 79912 | RNAseq | -0.8141 | 0.0009 | |
| TCGA | PYROXD1 | 79912 | RNAseq | -0.2256 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 2.
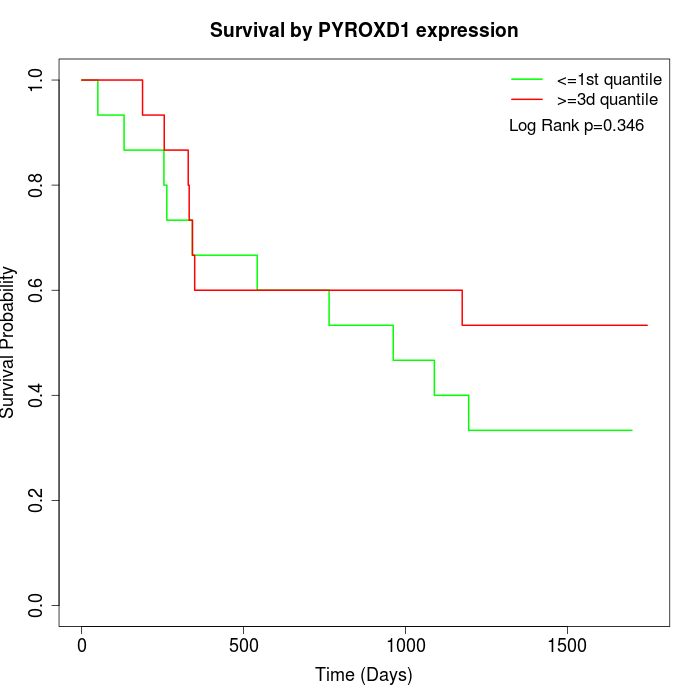
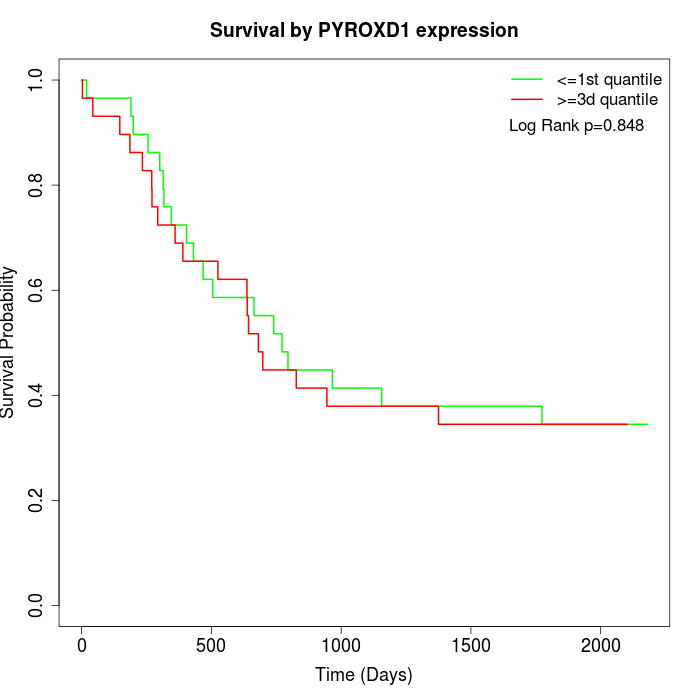
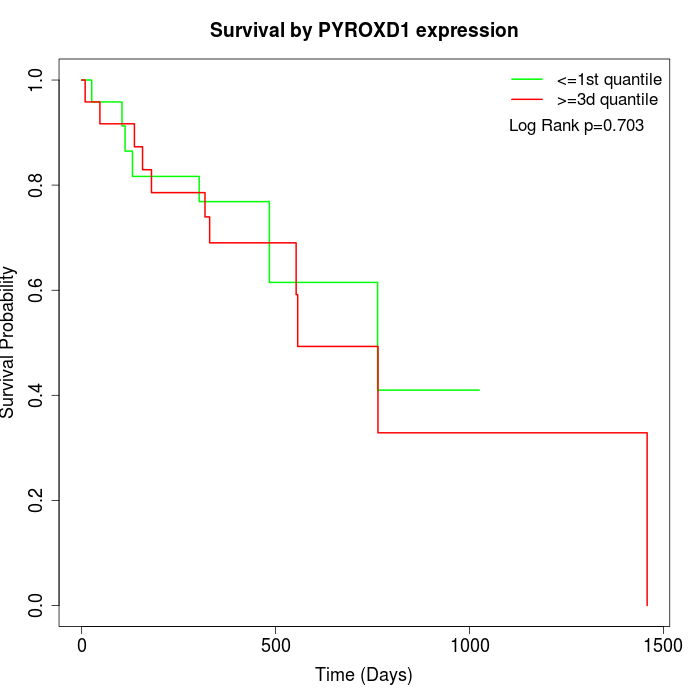
Survival by PYROXD1 expression:
Note: Click image to view full size file.
Copy number change of PYROXD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYROXD1 | 79912 | 9 | 2 | 19 | |
| GSE20123 | PYROXD1 | 79912 | 9 | 2 | 19 | |
| GSE43470 | PYROXD1 | 79912 | 9 | 4 | 30 | |
| GSE46452 | PYROXD1 | 79912 | 10 | 1 | 48 | |
| GSE47630 | PYROXD1 | 79912 | 14 | 1 | 25 | |
| GSE54993 | PYROXD1 | 79912 | 1 | 9 | 60 | |
| GSE54994 | PYROXD1 | 79912 | 11 | 2 | 40 | |
| GSE60625 | PYROXD1 | 79912 | 0 | 1 | 10 | |
| GSE74703 | PYROXD1 | 79912 | 9 | 3 | 24 | |
| GSE74704 | PYROXD1 | 79912 | 6 | 1 | 13 | |
| TCGA | PYROXD1 | 79912 | 42 | 5 | 49 |
Total number of gains: 120; Total number of losses: 31; Total Number of normals: 337.
Somatic mutations of PYROXD1:
Generating mutation plots.
Highly correlated genes for PYROXD1:
Showing top 20/627 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYROXD1 | SIKE1 | 0.756013 | 3 | 0 | 3 |
| PYROXD1 | ZMYM4 | 0.738165 | 3 | 0 | 3 |
| PYROXD1 | HDHD2 | 0.729176 | 3 | 0 | 3 |
| PYROXD1 | EPB41L5 | 0.714819 | 3 | 0 | 3 |
| PYROXD1 | ARL5B | 0.703657 | 4 | 0 | 4 |
| PYROXD1 | CYP2C19 | 0.702223 | 5 | 0 | 4 |
| PYROXD1 | UQCR11 | 0.699442 | 5 | 0 | 5 |
| PYROXD1 | STRAP | 0.697458 | 3 | 0 | 3 |
| PYROXD1 | EPS8 | 0.69458 | 4 | 0 | 4 |
| PYROXD1 | MKLN1 | 0.686906 | 4 | 0 | 4 |
| PYROXD1 | RMND1 | 0.68066 | 3 | 0 | 3 |
| PYROXD1 | COMMD3 | 0.677758 | 4 | 0 | 4 |
| PYROXD1 | WWTR1 | 0.67454 | 3 | 0 | 3 |
| PYROXD1 | NCOR1 | 0.673785 | 4 | 0 | 3 |
| PYROXD1 | TLE2 | 0.67252 | 4 | 0 | 4 |
| PYROXD1 | KCNAB1 | 0.66827 | 5 | 0 | 5 |
| PYROXD1 | TMEM109 | 0.668037 | 4 | 0 | 3 |
| PYROXD1 | DNAJC21 | 0.667976 | 5 | 0 | 5 |
| PYROXD1 | MACROD2 | 0.667789 | 4 | 0 | 3 |
| PYROXD1 | BAG3 | 0.667392 | 4 | 0 | 3 |
For details and further investigation, click here