| Full name: pyroglutamylated RFamide peptide receptor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4q27 | ||
| Entrez ID: 84109 | HGNC ID: HGNC:15565 | Ensembl Gene: ENSG00000186867 | OMIM ID: 606925 |
| Drug and gene relationship at DGIdb | |||
Expression of QRFPR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | QRFPR | 84109 | 1555533_at | 0.4156 | 0.2854 | |
| GSE26886 | QRFPR | 84109 | 1555533_at | 0.1403 | 0.2253 | |
| GSE45670 | QRFPR | 84109 | 1555533_at | 0.4229 | 0.1404 | |
| GSE53622 | QRFPR | 84109 | 120480 | 1.7118 | 0.0000 | |
| GSE53624 | QRFPR | 84109 | 120480 | 2.1757 | 0.0000 | |
| GSE63941 | QRFPR | 84109 | 1555533_at | 0.1917 | 0.7871 | |
| GSE77861 | QRFPR | 84109 | 1555533_at | 0.2795 | 0.0408 | |
| SRP007169 | QRFPR | 84109 | RNAseq | 4.7998 | 0.0000 | |
| SRP133303 | QRFPR | 84109 | RNAseq | 1.8686 | 0.0006 | |
| SRP159526 | QRFPR | 84109 | RNAseq | 4.8304 | 0.0000 | |
| TCGA | QRFPR | 84109 | RNAseq | 2.7671 | 0.0000 |
Upregulated datasets: 6; Downregulated datasets: 0.
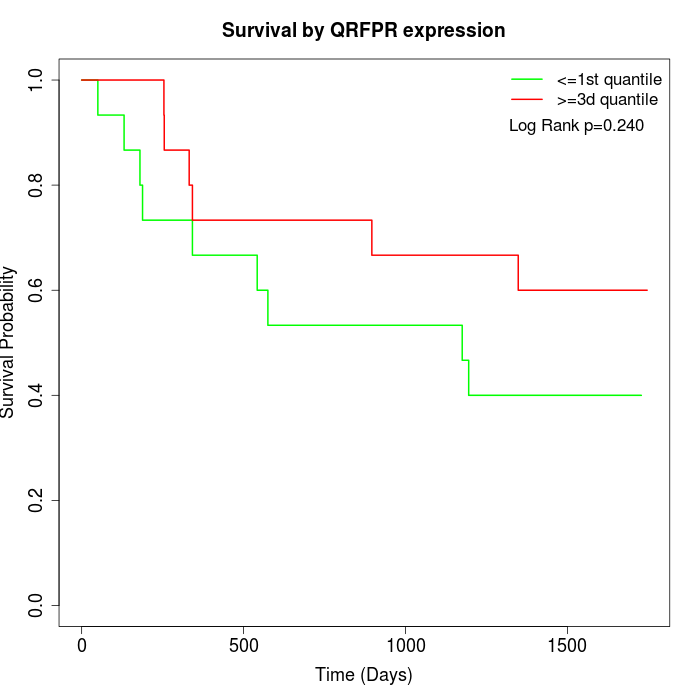
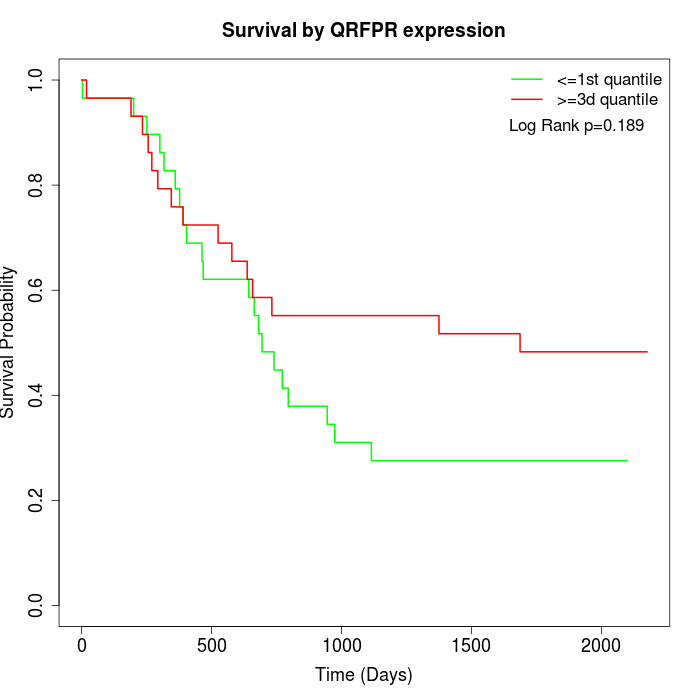
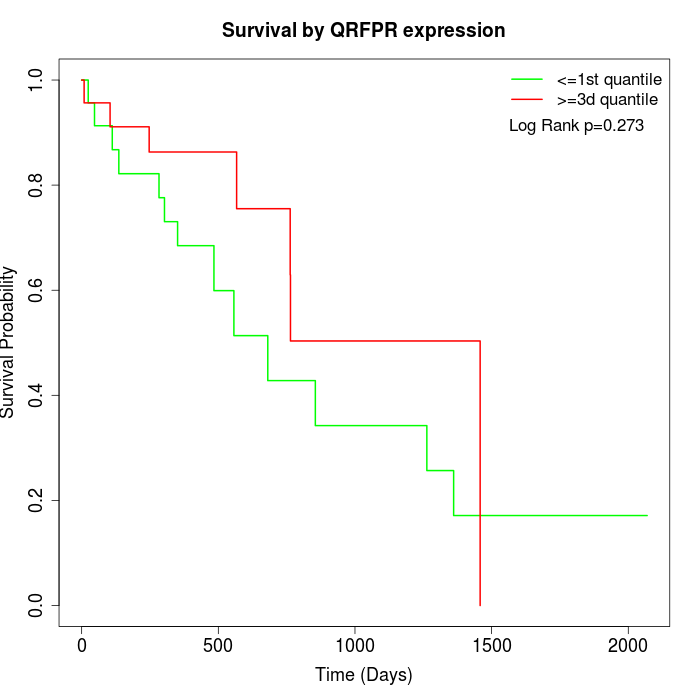
Survival by QRFPR expression:
Note: Click image to view full size file.
Copy number change of QRFPR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | QRFPR | 84109 | 0 | 13 | 17 | |
| GSE20123 | QRFPR | 84109 | 0 | 13 | 17 | |
| GSE43470 | QRFPR | 84109 | 0 | 13 | 30 | |
| GSE46452 | QRFPR | 84109 | 1 | 36 | 22 | |
| GSE47630 | QRFPR | 84109 | 0 | 22 | 18 | |
| GSE54993 | QRFPR | 84109 | 9 | 0 | 61 | |
| GSE54994 | QRFPR | 84109 | 1 | 10 | 42 | |
| GSE60625 | QRFPR | 84109 | 0 | 1 | 10 | |
| GSE74703 | QRFPR | 84109 | 0 | 11 | 25 | |
| GSE74704 | QRFPR | 84109 | 0 | 7 | 13 | |
| TCGA | QRFPR | 84109 | 8 | 34 | 54 |
Total number of gains: 19; Total number of losses: 160; Total Number of normals: 309.
Somatic mutations of QRFPR:
Generating mutation plots.
Highly correlated genes for QRFPR:
Showing top 20/134 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| QRFPR | CHRNA5 | 0.763668 | 4 | 0 | 4 |
| QRFPR | BCAM | 0.755347 | 3 | 0 | 3 |
| QRFPR | SULT1A1 | 0.745922 | 3 | 0 | 3 |
| QRFPR | ADCY3 | 0.71577 | 3 | 0 | 3 |
| QRFPR | TRMT61B | 0.706619 | 3 | 0 | 3 |
| QRFPR | RDH16 | 0.677622 | 3 | 0 | 3 |
| QRFPR | FOSL1 | 0.671604 | 3 | 0 | 3 |
| QRFPR | ARTN | 0.670932 | 4 | 0 | 4 |
| QRFPR | FAM169A | 0.665988 | 4 | 0 | 3 |
| QRFPR | CASC15 | 0.659258 | 4 | 0 | 4 |
| QRFPR | DMRT2 | 0.654855 | 6 | 0 | 6 |
| QRFPR | RPS6KA1 | 0.654542 | 3 | 0 | 3 |
| QRFPR | PAK1 | 0.654256 | 5 | 0 | 3 |
| QRFPR | KIF7 | 0.647748 | 3 | 0 | 3 |
| QRFPR | CHST12 | 0.636559 | 3 | 0 | 3 |
| QRFPR | RGS20 | 0.636291 | 3 | 0 | 3 |
| QRFPR | SCYL1 | 0.632377 | 3 | 0 | 3 |
| QRFPR | KREMEN1 | 0.63217 | 4 | 0 | 3 |
| QRFPR | CCDC85B | 0.629987 | 3 | 0 | 3 |
| QRFPR | SLC7A11 | 0.622729 | 6 | 0 | 6 |
For details and further investigation, click here