| Full name: Rap guanine nucleotide exchange factor 1 | Alias Symbol: C3G | ||
| Type: protein-coding gene | Cytoband: 9q34.13 | ||
| Entrez ID: 2889 | HGNC ID: HGNC:4568 | Ensembl Gene: ENSG00000107263 | OMIM ID: 600303 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
RAPGEF1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa04722 | Neurotrophin signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa05211 | Renal cell carcinoma |
Expression of RAPGEF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAPGEF1 | 2889 | 226389_s_at | 0.3689 | 0.4051 | |
| GSE20347 | RAPGEF1 | 2889 | 204543_at | 0.0120 | 0.9002 | |
| GSE23400 | RAPGEF1 | 2889 | 204543_at | -0.0854 | 0.0152 | |
| GSE26886 | RAPGEF1 | 2889 | 226389_s_at | 0.2923 | 0.0564 | |
| GSE29001 | RAPGEF1 | 2889 | 204543_at | -0.0739 | 0.6367 | |
| GSE38129 | RAPGEF1 | 2889 | 204543_at | -0.1589 | 0.0186 | |
| GSE45670 | RAPGEF1 | 2889 | 226389_s_at | 0.0842 | 0.3943 | |
| GSE53622 | RAPGEF1 | 2889 | 129350 | -0.4844 | 0.0002 | |
| GSE53624 | RAPGEF1 | 2889 | 129350 | -0.9970 | 0.0000 | |
| GSE63941 | RAPGEF1 | 2889 | 225738_at | 0.2085 | 0.6358 | |
| GSE77861 | RAPGEF1 | 2889 | 226389_s_at | 0.2423 | 0.0790 | |
| GSE97050 | RAPGEF1 | 2889 | A_23_P391764 | 0.1105 | 0.6226 | |
| SRP007169 | RAPGEF1 | 2889 | RNAseq | 1.8390 | 0.0000 | |
| SRP008496 | RAPGEF1 | 2889 | RNAseq | 1.4839 | 0.0000 | |
| SRP064894 | RAPGEF1 | 2889 | RNAseq | 0.5025 | 0.0032 | |
| SRP133303 | RAPGEF1 | 2889 | RNAseq | 0.4773 | 0.0072 | |
| SRP159526 | RAPGEF1 | 2889 | RNAseq | 0.4361 | 0.1919 | |
| SRP193095 | RAPGEF1 | 2889 | RNAseq | 0.6586 | 0.0000 | |
| SRP219564 | RAPGEF1 | 2889 | RNAseq | 0.2491 | 0.4722 | |
| TCGA | RAPGEF1 | 2889 | RNAseq | -0.0735 | 0.1585 |
Upregulated datasets: 2; Downregulated datasets: 0.
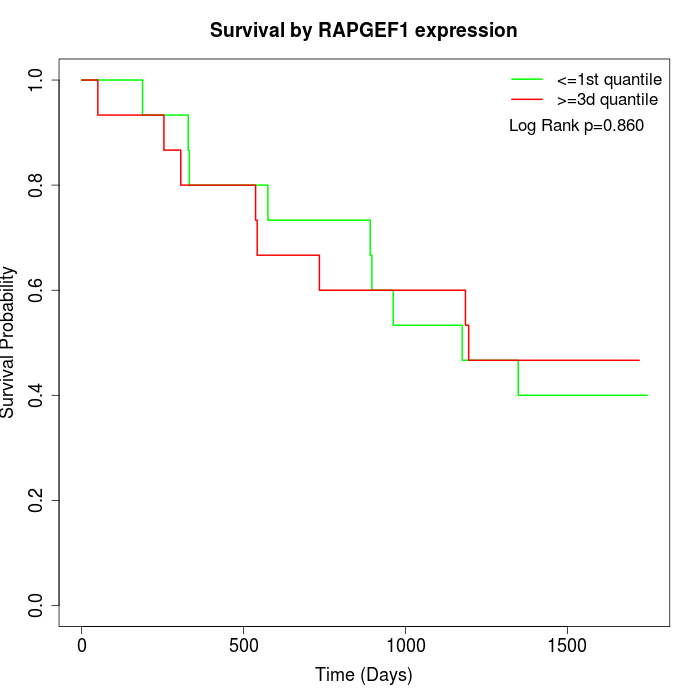
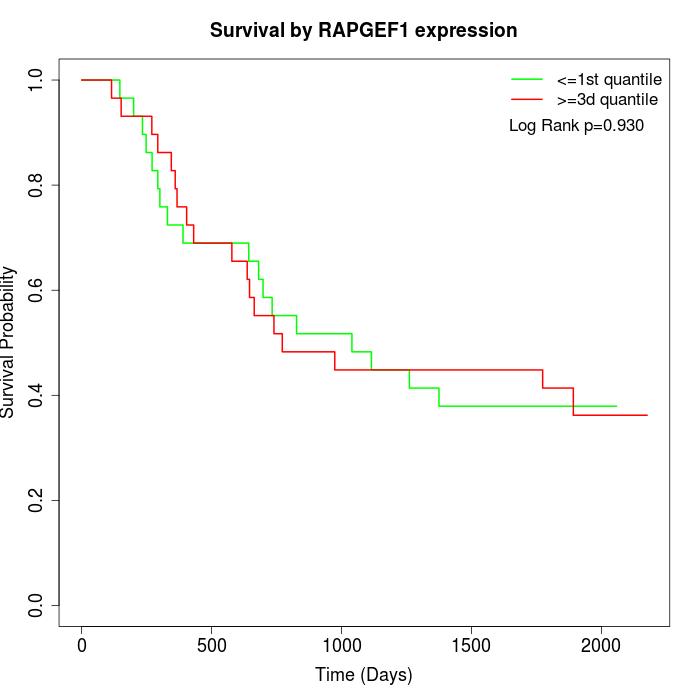
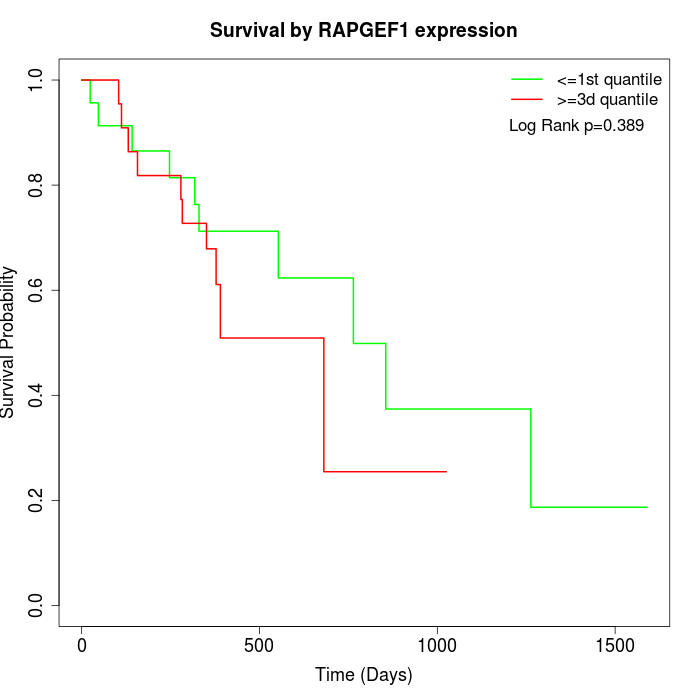
Survival by RAPGEF1 expression:
Note: Click image to view full size file.
Copy number change of RAPGEF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAPGEF1 | 2889 | 6 | 9 | 15 | |
| GSE20123 | RAPGEF1 | 2889 | 6 | 9 | 15 | |
| GSE43470 | RAPGEF1 | 2889 | 5 | 7 | 31 | |
| GSE46452 | RAPGEF1 | 2889 | 6 | 13 | 40 | |
| GSE47630 | RAPGEF1 | 2889 | 4 | 15 | 21 | |
| GSE54993 | RAPGEF1 | 2889 | 3 | 3 | 64 | |
| GSE54994 | RAPGEF1 | 2889 | 12 | 9 | 32 | |
| GSE60625 | RAPGEF1 | 2889 | 0 | 0 | 11 | |
| GSE74703 | RAPGEF1 | 2889 | 5 | 5 | 26 | |
| GSE74704 | RAPGEF1 | 2889 | 3 | 7 | 10 | |
| TCGA | RAPGEF1 | 2889 | 28 | 26 | 42 |
Total number of gains: 78; Total number of losses: 103; Total Number of normals: 307.
Somatic mutations of RAPGEF1:
Generating mutation plots.
Highly correlated genes for RAPGEF1:
Showing top 20/312 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAPGEF1 | STAT5B | 0.807267 | 3 | 0 | 3 |
| RAPGEF1 | SIDT2 | 0.782636 | 3 | 0 | 3 |
| RAPGEF1 | CLDN18 | 0.77381 | 3 | 0 | 3 |
| RAPGEF1 | ZMIZ2 | 0.759537 | 3 | 0 | 3 |
| RAPGEF1 | KIR3DL2 | 0.756938 | 3 | 0 | 3 |
| RAPGEF1 | SEMA6A | 0.753218 | 3 | 0 | 3 |
| RAPGEF1 | SZT2 | 0.748759 | 3 | 0 | 3 |
| RAPGEF1 | BSN | 0.746501 | 3 | 0 | 3 |
| RAPGEF1 | NLE1 | 0.745283 | 3 | 0 | 3 |
| RAPGEF1 | CLIP3 | 0.737588 | 4 | 0 | 4 |
| RAPGEF1 | APBB1IP | 0.73706 | 3 | 0 | 3 |
| RAPGEF1 | TRAF7 | 0.736319 | 3 | 0 | 3 |
| RAPGEF1 | CLDN11 | 0.734833 | 4 | 0 | 4 |
| RAPGEF1 | GOLGA8A | 0.732554 | 3 | 0 | 3 |
| RAPGEF1 | NIPSNAP3B | 0.730059 | 3 | 0 | 3 |
| RAPGEF1 | COQ10B | 0.72099 | 3 | 0 | 3 |
| RAPGEF1 | IL3RA | 0.715347 | 3 | 0 | 3 |
| RAPGEF1 | GDF5 | 0.715117 | 4 | 0 | 4 |
| RAPGEF1 | FAM160B1 | 0.710285 | 3 | 0 | 3 |
| RAPGEF1 | SLC39A3 | 0.709922 | 3 | 0 | 3 |
For details and further investigation, click here