| Full name: regulator of calcineurin 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 21q22.12 | ||
| Entrez ID: 1827 | HGNC ID: HGNC:3040 | Ensembl Gene: ENSG00000159200 | OMIM ID: 602917 |
| Drug and gene relationship at DGIdb | |||
RCAN1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04919 | Thyroid hormone signaling pathway | |
| hsa04921 | Oxytocin signaling pathway |
Expression of RCAN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RCAN1 | 1827 | 208370_s_at | 0.3610 | 0.3039 | |
| GSE20347 | RCAN1 | 1827 | 208370_s_at | 0.4417 | 0.0806 | |
| GSE23400 | RCAN1 | 1827 | 208370_s_at | 0.3316 | 0.1080 | |
| GSE26886 | RCAN1 | 1827 | 208370_s_at | -0.2209 | 0.5068 | |
| GSE29001 | RCAN1 | 1827 | 208370_s_at | 0.7505 | 0.0276 | |
| GSE38129 | RCAN1 | 1827 | 208370_s_at | 0.2393 | 0.3976 | |
| GSE45670 | RCAN1 | 1827 | 208370_s_at | 0.1425 | 0.6578 | |
| GSE53622 | RCAN1 | 1827 | 21259 | 0.2182 | 0.1904 | |
| GSE53624 | RCAN1 | 1827 | 21259 | 0.6388 | 0.0000 | |
| GSE63941 | RCAN1 | 1827 | 208370_s_at | -3.1948 | 0.0005 | |
| GSE77861 | RCAN1 | 1827 | 208370_s_at | 0.8043 | 0.1465 | |
| GSE97050 | RCAN1 | 1827 | A_23_P166248 | -0.1722 | 0.7400 | |
| SRP007169 | RCAN1 | 1827 | RNAseq | 2.5159 | 0.0000 | |
| SRP008496 | RCAN1 | 1827 | RNAseq | 2.0171 | 0.0000 | |
| SRP064894 | RCAN1 | 1827 | RNAseq | 0.6311 | 0.0627 | |
| SRP133303 | RCAN1 | 1827 | RNAseq | 0.4293 | 0.1382 | |
| SRP159526 | RCAN1 | 1827 | RNAseq | 0.8334 | 0.0542 | |
| SRP193095 | RCAN1 | 1827 | RNAseq | 1.1647 | 0.0000 | |
| SRP219564 | RCAN1 | 1827 | RNAseq | -0.3387 | 0.6718 | |
| TCGA | RCAN1 | 1827 | RNAseq | -0.1776 | 0.0419 |
Upregulated datasets: 3; Downregulated datasets: 1.
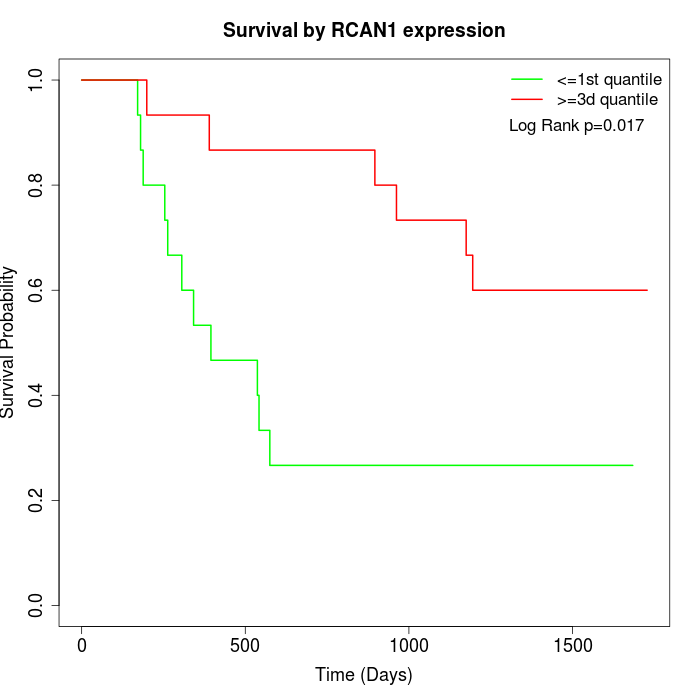
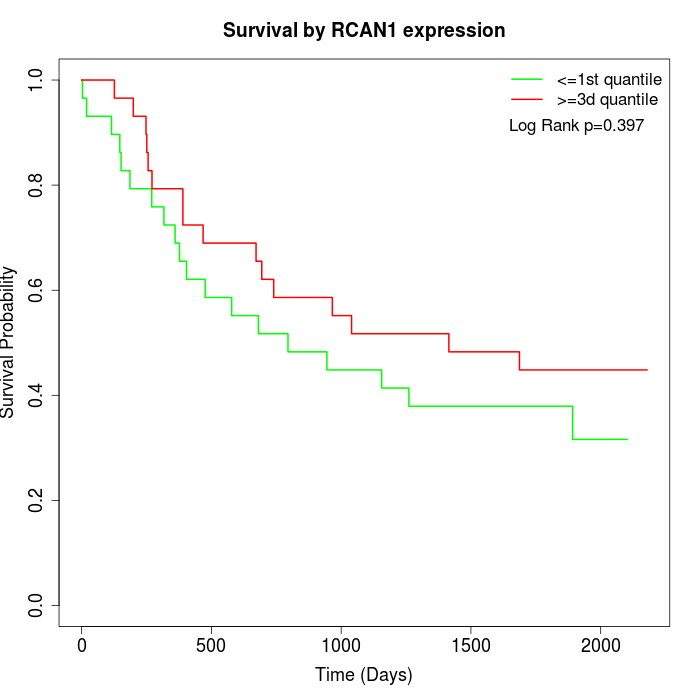
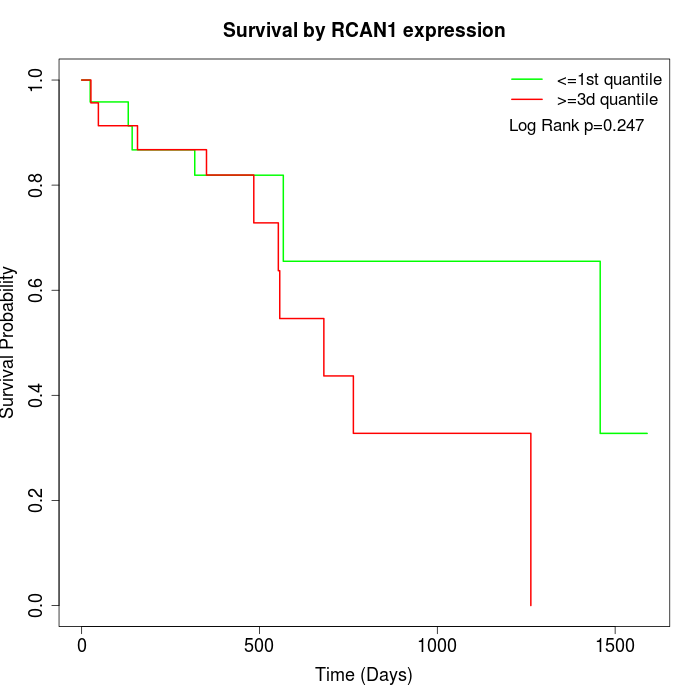
Survival by RCAN1 expression:
Note: Click image to view full size file.
Copy number change of RCAN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RCAN1 | 1827 | 2 | 7 | 21 | |
| GSE20123 | RCAN1 | 1827 | 2 | 8 | 20 | |
| GSE43470 | RCAN1 | 1827 | 1 | 11 | 31 | |
| GSE46452 | RCAN1 | 1827 | 1 | 21 | 37 | |
| GSE47630 | RCAN1 | 1827 | 6 | 17 | 17 | |
| GSE54993 | RCAN1 | 1827 | 9 | 1 | 60 | |
| GSE54994 | RCAN1 | 1827 | 1 | 9 | 43 | |
| GSE60625 | RCAN1 | 1827 | 0 | 0 | 11 | |
| GSE74703 | RCAN1 | 1827 | 1 | 8 | 27 | |
| GSE74704 | RCAN1 | 1827 | 1 | 4 | 15 | |
| TCGA | RCAN1 | 1827 | 8 | 37 | 51 |
Total number of gains: 32; Total number of losses: 123; Total Number of normals: 333.
Somatic mutations of RCAN1:
Generating mutation plots.
Highly correlated genes for RCAN1:
Showing top 20/727 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RCAN1 | ATL1 | 0.811683 | 3 | 0 | 3 |
| RCAN1 | FZD4 | 0.765999 | 3 | 0 | 3 |
| RCAN1 | RTF1 | 0.765099 | 3 | 0 | 3 |
| RCAN1 | SGTB | 0.751296 | 3 | 0 | 3 |
| RCAN1 | LETM2 | 0.734736 | 3 | 0 | 3 |
| RCAN1 | RCN2 | 0.732278 | 3 | 0 | 3 |
| RCAN1 | CNTNAP1 | 0.730877 | 3 | 0 | 3 |
| RCAN1 | KBTBD6 | 0.727713 | 4 | 0 | 4 |
| RCAN1 | DZIP1L | 0.722014 | 3 | 0 | 3 |
| RCAN1 | DHX57 | 0.717207 | 3 | 0 | 3 |
| RCAN1 | PACSIN2 | 0.70665 | 3 | 0 | 3 |
| RCAN1 | TMOD2 | 0.704546 | 4 | 0 | 3 |
| RCAN1 | FILIP1L | 0.699128 | 9 | 0 | 8 |
| RCAN1 | PLEKHM3 | 0.697469 | 3 | 0 | 3 |
| RCAN1 | MOB3C | 0.692452 | 3 | 0 | 3 |
| RCAN1 | PRICKLE1 | 0.692224 | 4 | 0 | 4 |
| RCAN1 | IWS1 | 0.689674 | 3 | 0 | 3 |
| RCAN1 | DPY19L3 | 0.688246 | 3 | 0 | 3 |
| RCAN1 | TRPC4AP | 0.684921 | 3 | 0 | 3 |
| RCAN1 | MAGOHB | 0.680234 | 3 | 0 | 3 |
For details and further investigation, click here