| Full name: retinal degeneration 3, GUCY2D regulator | Alias Symbol: LCA12 | ||
| Type: protein-coding gene | Cytoband: 1q32.3 | ||
| Entrez ID: 343035 | HGNC ID: HGNC:19689 | Ensembl Gene: ENSG00000198570 | OMIM ID: 180040 |
| Drug and gene relationship at DGIdb | |||
Expression of RD3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RD3 | 343035 | 236563_at | -0.2294 | 0.3120 | |
| GSE26886 | RD3 | 343035 | 236563_at | -0.0932 | 0.2212 | |
| GSE45670 | RD3 | 343035 | 236563_at | 0.0736 | 0.3660 | |
| GSE53622 | RD3 | 343035 | 74131 | -0.2279 | 0.2508 | |
| GSE53624 | RD3 | 343035 | 74131 | -0.7256 | 0.0002 | |
| GSE63941 | RD3 | 343035 | 236563_at | 0.0033 | 0.9837 | |
| GSE77861 | RD3 | 343035 | 236563_at | -0.0900 | 0.2144 | |
| SRP133303 | RD3 | 343035 | RNAseq | 0.3784 | 0.1073 | |
| SRP193095 | RD3 | 343035 | RNAseq | -0.1144 | 0.5458 | |
| TCGA | RD3 | 343035 | RNAseq | -3.1301 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
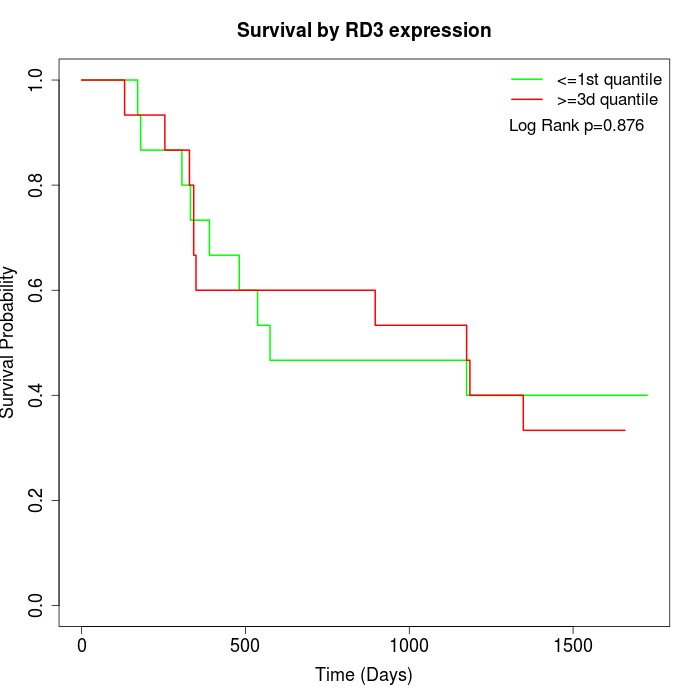
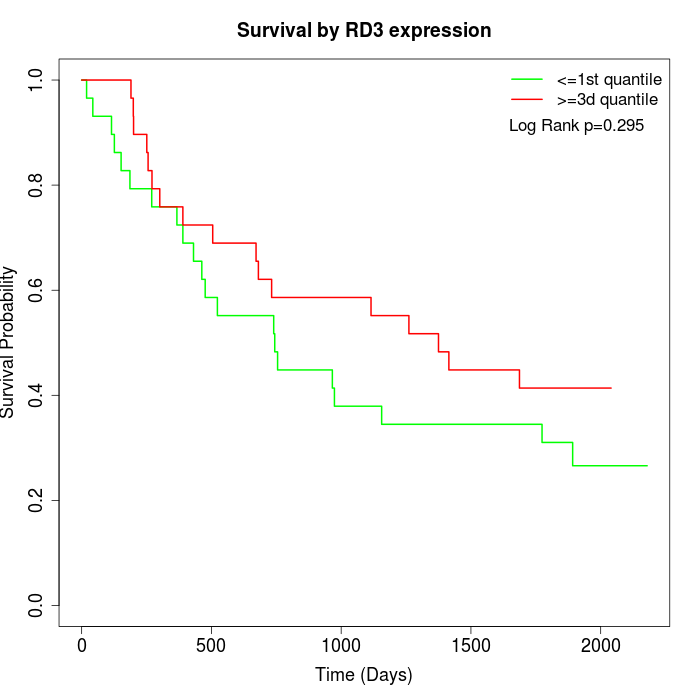
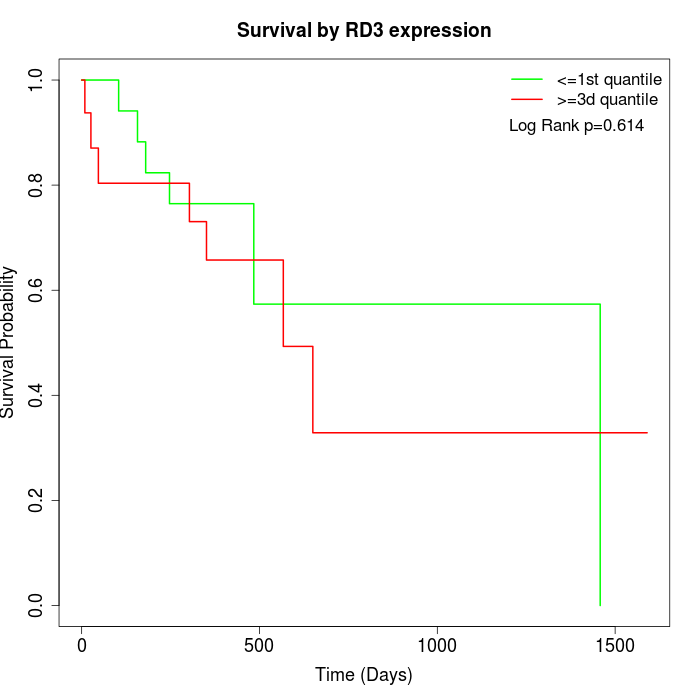
Survival by RD3 expression:
Note: Click image to view full size file.
Copy number change of RD3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RD3 | 343035 | 11 | 0 | 19 | |
| GSE20123 | RD3 | 343035 | 10 | 0 | 20 | |
| GSE43470 | RD3 | 343035 | 7 | 0 | 36 | |
| GSE46452 | RD3 | 343035 | 3 | 1 | 55 | |
| GSE47630 | RD3 | 343035 | 15 | 0 | 25 | |
| GSE54993 | RD3 | 343035 | 0 | 6 | 64 | |
| GSE54994 | RD3 | 343035 | 13 | 0 | 40 | |
| GSE60625 | RD3 | 343035 | 0 | 0 | 11 | |
| GSE74703 | RD3 | 343035 | 7 | 0 | 29 | |
| GSE74704 | RD3 | 343035 | 4 | 0 | 16 | |
| TCGA | RD3 | 343035 | 45 | 3 | 48 |
Total number of gains: 115; Total number of losses: 10; Total Number of normals: 363.
Somatic mutations of RD3:
Generating mutation plots.
Highly correlated genes for RD3:
Showing top 20/73 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RD3 | NAALADL1 | 0.697348 | 3 | 0 | 3 |
| RD3 | TPSD1 | 0.680009 | 3 | 0 | 3 |
| RD3 | GRHPR | 0.672566 | 3 | 0 | 3 |
| RD3 | EGFLAM-AS2 | 0.660262 | 3 | 0 | 3 |
| RD3 | COL14A1 | 0.643674 | 3 | 0 | 3 |
| RD3 | PRSS33 | 0.635533 | 3 | 0 | 3 |
| RD3 | FMN1 | 0.633034 | 3 | 0 | 3 |
| RD3 | ADH1B | 0.629583 | 3 | 0 | 3 |
| RD3 | RELN | 0.629544 | 3 | 0 | 3 |
| RD3 | SLCO2B1 | 0.628848 | 3 | 0 | 3 |
| RD3 | NME5 | 0.626941 | 3 | 0 | 3 |
| RD3 | KLK4 | 0.626675 | 5 | 0 | 5 |
| RD3 | TCL6 | 0.626545 | 3 | 0 | 3 |
| RD3 | HTR1E | 0.621838 | 3 | 0 | 3 |
| RD3 | SH3BGRL | 0.620522 | 3 | 0 | 3 |
| RD3 | S100B | 0.613107 | 4 | 0 | 4 |
| RD3 | ITM2A | 0.611178 | 4 | 0 | 3 |
| RD3 | LINC00589 | 0.610681 | 3 | 0 | 3 |
| RD3 | EBF3 | 0.610374 | 3 | 0 | 3 |
| RD3 | ARHGEF6 | 0.607344 | 3 | 0 | 3 |
For details and further investigation, click here