| Full name: ral guanine nucleotide dissociation stimulator like 3 | Alias Symbol: FLJ32585 | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 57139 | HGNC ID: HGNC:30282 | Ensembl Gene: ENSG00000205517 | OMIM ID: 616743 |
| Drug and gene relationship at DGIdb | |||
Expression of RGL3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGL3 | 57139 | 1556355_x_at | 0.0257 | 0.9455 | |
| GSE26886 | RGL3 | 57139 | 228877_at | 0.1555 | 0.1333 | |
| GSE45670 | RGL3 | 57139 | 1556355_x_at | 0.0716 | 0.4371 | |
| GSE53622 | RGL3 | 57139 | 45104 | -0.4643 | 0.0211 | |
| GSE53624 | RGL3 | 57139 | 45104 | -0.6135 | 0.0000 | |
| GSE63941 | RGL3 | 57139 | 1556355_x_at | 0.4791 | 0.0033 | |
| GSE77861 | RGL3 | 57139 | 228877_at | -0.0910 | 0.4071 | |
| GSE97050 | RGL3 | 57139 | A_33_P3293213 | 0.2247 | 0.5036 | |
| SRP064894 | RGL3 | 57139 | RNAseq | -0.2174 | 0.4689 | |
| SRP133303 | RGL3 | 57139 | RNAseq | -0.4802 | 0.4035 | |
| SRP219564 | RGL3 | 57139 | RNAseq | 1.2984 | 0.0780 | |
| TCGA | RGL3 | 57139 | RNAseq | -2.2587 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
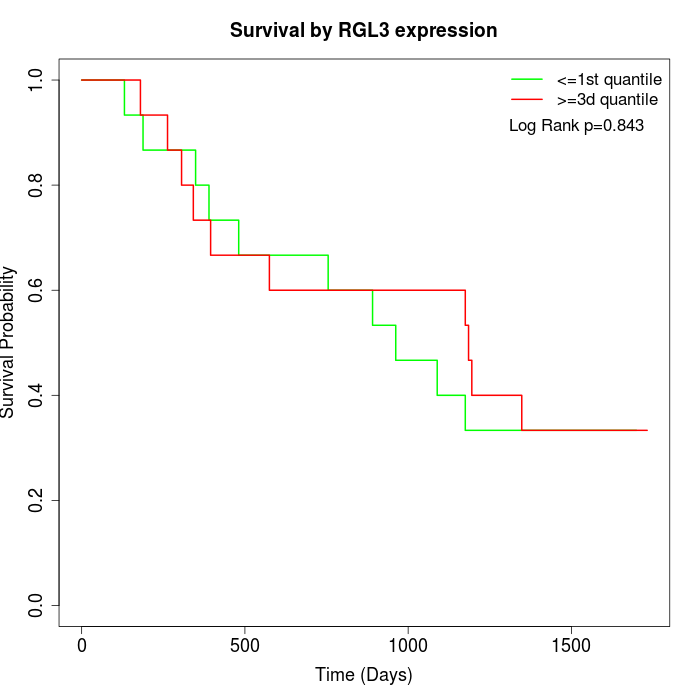
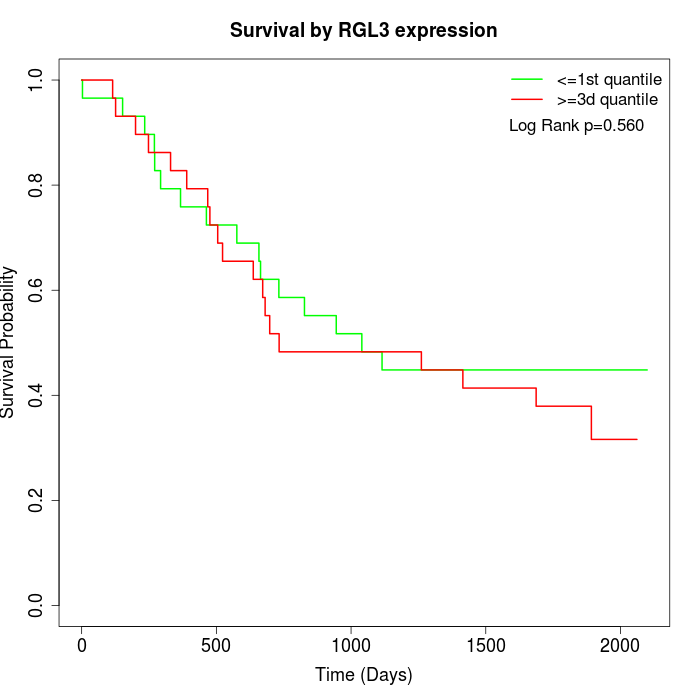
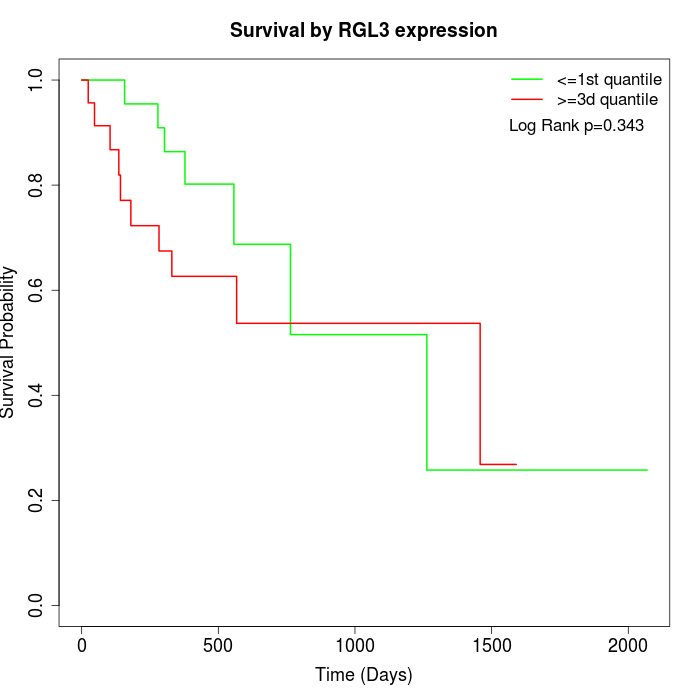
Survival by RGL3 expression:
Note: Click image to view full size file.
Copy number change of RGL3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGL3 | 57139 | 4 | 2 | 24 | |
| GSE20123 | RGL3 | 57139 | 3 | 1 | 26 | |
| GSE43470 | RGL3 | 57139 | 2 | 6 | 35 | |
| GSE46452 | RGL3 | 57139 | 47 | 1 | 11 | |
| GSE47630 | RGL3 | 57139 | 4 | 7 | 29 | |
| GSE54993 | RGL3 | 57139 | 16 | 3 | 51 | |
| GSE54994 | RGL3 | 57139 | 6 | 13 | 34 | |
| GSE60625 | RGL3 | 57139 | 9 | 0 | 2 | |
| GSE74703 | RGL3 | 57139 | 2 | 4 | 30 | |
| GSE74704 | RGL3 | 57139 | 0 | 1 | 19 | |
| TCGA | RGL3 | 57139 | 16 | 12 | 68 |
Total number of gains: 109; Total number of losses: 50; Total Number of normals: 329.
Somatic mutations of RGL3:
Generating mutation plots.
Highly correlated genes for RGL3:
Showing top 20/32 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGL3 | SFN | 0.665429 | 3 | 0 | 3 |
| RGL3 | TNNT1 | 0.664496 | 3 | 0 | 3 |
| RGL3 | CENPT | 0.65939 | 3 | 0 | 3 |
| RGL3 | NCR2 | 0.632132 | 4 | 0 | 4 |
| RGL3 | SPEM1 | 0.625943 | 3 | 0 | 3 |
| RGL3 | DPPA4 | 0.623463 | 3 | 0 | 3 |
| RGL3 | CSF2 | 0.606802 | 4 | 0 | 3 |
| RGL3 | PRM2 | 0.605006 | 4 | 0 | 3 |
| RGL3 | NEUROG2 | 0.598022 | 3 | 0 | 3 |
| RGL3 | TM6SF2 | 0.59595 | 3 | 0 | 3 |
| RGL3 | CLDN9 | 0.589321 | 3 | 0 | 3 |
| RGL3 | FLT4 | 0.588368 | 4 | 0 | 3 |
| RGL3 | SLC19A3 | 0.582656 | 3 | 0 | 3 |
| RGL3 | CA7 | 0.582047 | 4 | 0 | 3 |
| RGL3 | CPNE7 | 0.581147 | 4 | 0 | 3 |
| RGL3 | CBLC | 0.579741 | 4 | 0 | 3 |
| RGL3 | SLC22A13 | 0.578622 | 4 | 0 | 4 |
| RGL3 | MAP4K2 | 0.563278 | 3 | 0 | 3 |
| RGL3 | GPR52 | 0.562104 | 3 | 0 | 3 |
| RGL3 | KCNJ13 | 0.555713 | 5 | 0 | 3 |
For details and further investigation, click here