| Full name: regulator of G protein signaling 22 | Alias Symbol: DKFZP434I092|PRTD-NY2|CT145 | ||
| Type: protein-coding gene | Cytoband: 8q22.2 | ||
| Entrez ID: 26166 | HGNC ID: HGNC:24499 | Ensembl Gene: ENSG00000132554 | OMIM ID: 615650 |
| Drug and gene relationship at DGIdb | |||
Expression of RGS22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGS22 | 26166 | 223691_at | -0.1688 | 0.4593 | |
| GSE26886 | RGS22 | 26166 | 223691_at | 0.1878 | 0.2663 | |
| GSE45670 | RGS22 | 26166 | 223691_at | -0.4180 | 0.0022 | |
| GSE53622 | RGS22 | 26166 | 7717 | -0.5718 | 0.0000 | |
| GSE53624 | RGS22 | 26166 | 7717 | -1.0227 | 0.0000 | |
| GSE63941 | RGS22 | 26166 | 223691_at | -0.3142 | 0.2593 | |
| GSE77861 | RGS22 | 26166 | 223691_at | -0.0003 | 0.9980 | |
| GSE97050 | RGS22 | 26166 | A_32_P125771 | 0.0893 | 0.6701 | |
| SRP133303 | RGS22 | 26166 | RNAseq | -2.3713 | 0.0000 | |
| SRP159526 | RGS22 | 26166 | RNAseq | -0.9627 | 0.0573 | |
| TCGA | RGS22 | 26166 | RNAseq | -1.7321 | 0.0131 |
Upregulated datasets: 0; Downregulated datasets: 3.
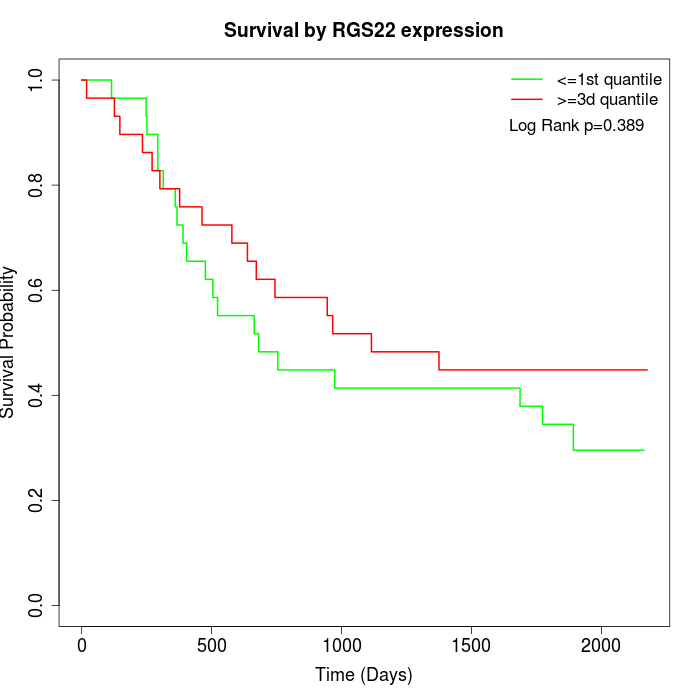
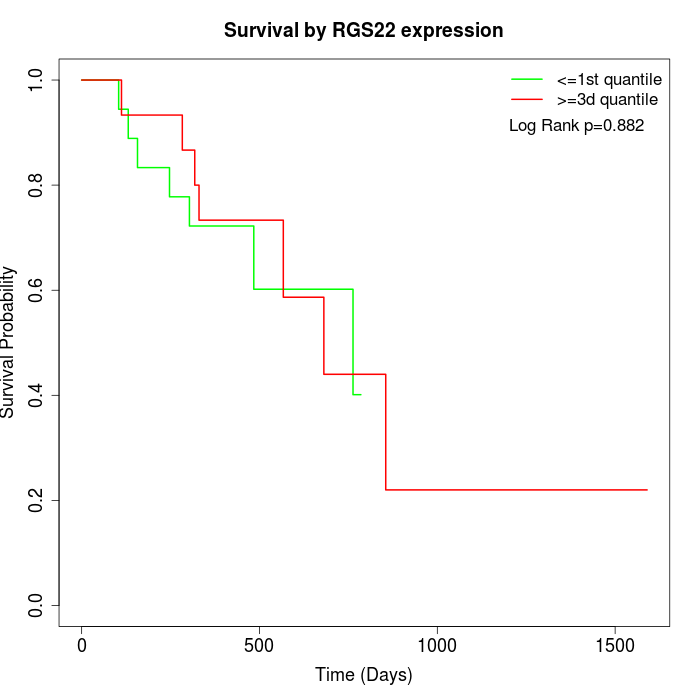
Survival by RGS22 expression:
Note: Click image to view full size file.
Copy number change of RGS22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGS22 | 26166 | 17 | 0 | 13 | |
| GSE20123 | RGS22 | 26166 | 17 | 0 | 13 | |
| GSE43470 | RGS22 | 26166 | 21 | 0 | 22 | |
| GSE46452 | RGS22 | 26166 | 24 | 0 | 35 | |
| GSE47630 | RGS22 | 26166 | 24 | 0 | 16 | |
| GSE54993 | RGS22 | 26166 | 0 | 20 | 50 | |
| GSE54994 | RGS22 | 26166 | 38 | 1 | 14 | |
| GSE60625 | RGS22 | 26166 | 0 | 4 | 7 | |
| GSE74703 | RGS22 | 26166 | 18 | 0 | 18 | |
| GSE74704 | RGS22 | 26166 | 12 | 0 | 8 | |
| TCGA | RGS22 | 26166 | 58 | 1 | 37 |
Total number of gains: 229; Total number of losses: 26; Total Number of normals: 233.
Somatic mutations of RGS22:
Generating mutation plots.
Highly correlated genes for RGS22:
Showing top 20/93 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGS22 | FIGLA | 0.740722 | 3 | 0 | 3 |
| RGS22 | SDCBP2 | 0.663469 | 3 | 0 | 3 |
| RGS22 | HAMP | 0.648866 | 3 | 0 | 3 |
| RGS22 | DEGS2 | 0.644413 | 3 | 0 | 3 |
| RGS22 | MUC2 | 0.641581 | 3 | 0 | 3 |
| RGS22 | RAET1E | 0.639683 | 3 | 0 | 3 |
| RGS22 | SPRR2A | 0.63707 | 3 | 0 | 3 |
| RGS22 | SLC34A3 | 0.636381 | 3 | 0 | 3 |
| RGS22 | TGM5 | 0.629327 | 3 | 0 | 3 |
| RGS22 | LIPM | 0.624628 | 3 | 0 | 3 |
| RGS22 | SLC10A6 | 0.623202 | 3 | 0 | 3 |
| RGS22 | IL17B | 0.623014 | 3 | 0 | 3 |
| RGS22 | SPRR2E | 0.62196 | 3 | 0 | 3 |
| RGS22 | RNF222 | 0.619875 | 3 | 0 | 3 |
| RGS22 | CIITA | 0.61956 | 3 | 0 | 3 |
| RGS22 | MAST3 | 0.618906 | 3 | 0 | 3 |
| RGS22 | EFNA5 | 0.61284 | 3 | 0 | 3 |
| RGS22 | SPRR1A | 0.61072 | 3 | 0 | 3 |
| RGS22 | SPRR2D | 0.610066 | 3 | 0 | 3 |
| RGS22 | DCD | 0.607962 | 4 | 0 | 4 |
For details and further investigation, click here