| Full name: regulator of G protein signaling 9 binding protein | Alias Symbol: FLJ45744|PERRS|R9AP|RGS9 | ||
| Type: protein-coding gene | Cytoband: 19q13.11 | ||
| Entrez ID: 388531 | HGNC ID: HGNC:30304 | Ensembl Gene: ENSG00000186326 | OMIM ID: 607814 |
| Drug and gene relationship at DGIdb | |||
Expression of RGS9BP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGS9BP | 388531 | 1556199_a_at | -0.1523 | 0.5443 | |
| GSE26886 | RGS9BP | 388531 | 1556199_a_at | -0.0991 | 0.5678 | |
| GSE45670 | RGS9BP | 388531 | 1556199_a_at | -0.1205 | 0.3749 | |
| GSE53622 | RGS9BP | 388531 | 141537 | -2.0742 | 0.0000 | |
| GSE53624 | RGS9BP | 388531 | 141537 | -1.7767 | 0.0000 | |
| GSE63941 | RGS9BP | 388531 | 1556199_a_at | 0.1335 | 0.3650 | |
| GSE77861 | RGS9BP | 388531 | 1556199_a_at | -0.1465 | 0.2361 | |
| GSE97050 | RGS9BP | 388531 | A_32_P227605 | -0.4883 | 0.2018 | |
| SRP133303 | RGS9BP | 388531 | RNAseq | -1.5840 | 0.0154 | |
| TCGA | RGS9BP | 388531 | RNAseq | -0.6289 | 0.0667 |
Upregulated datasets: 0; Downregulated datasets: 3.
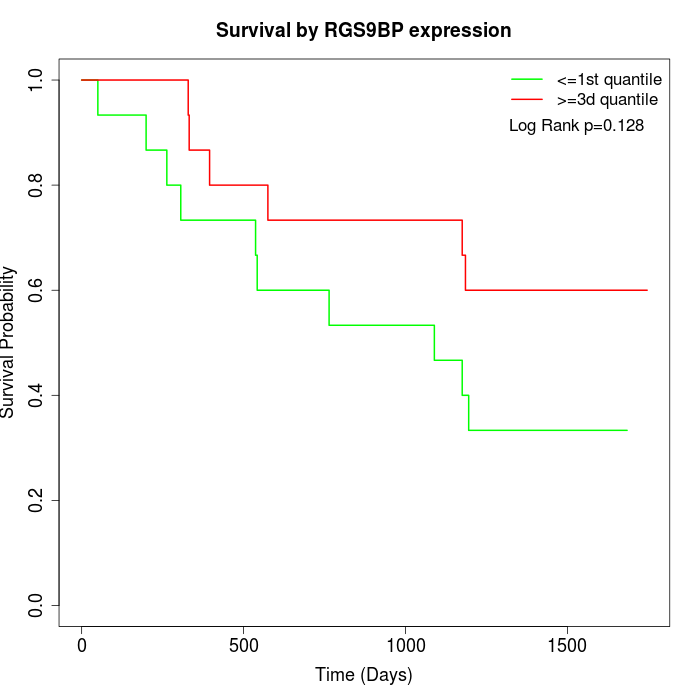
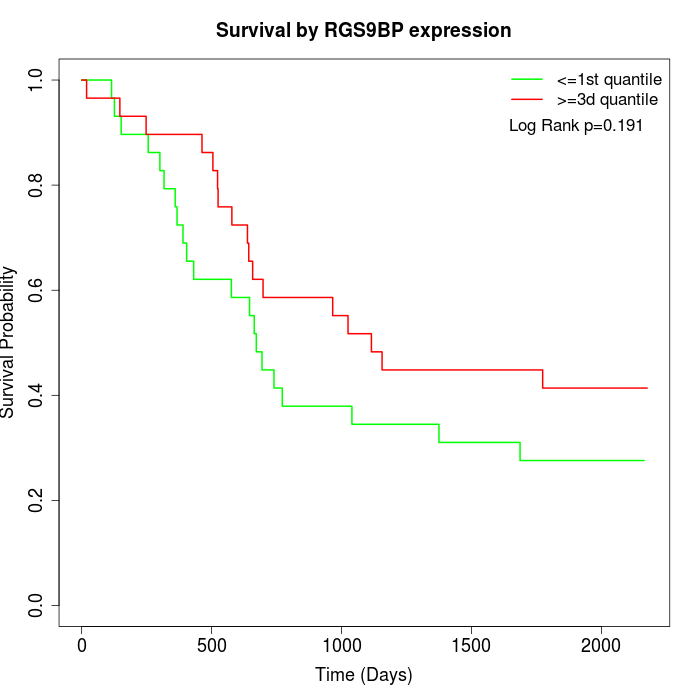
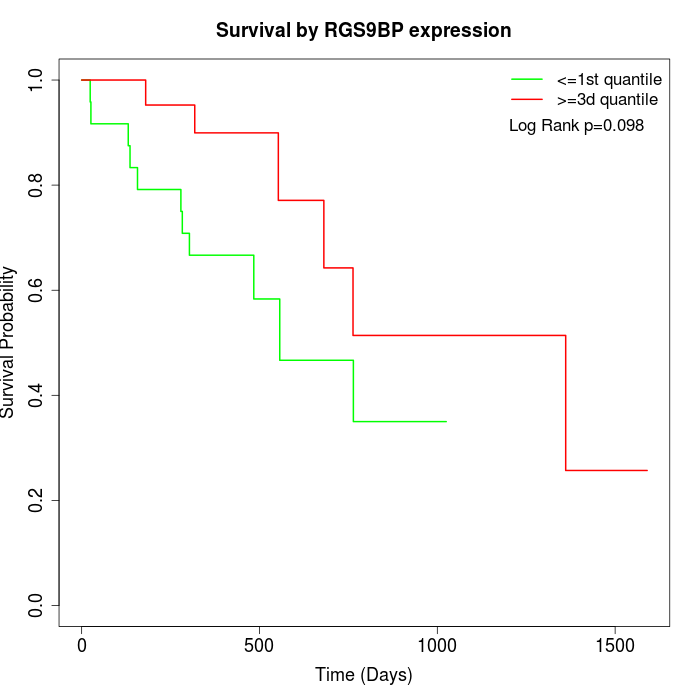
Survival by RGS9BP expression:
Note: Click image to view full size file.
Copy number change of RGS9BP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGS9BP | 388531 | 7 | 6 | 17 | |
| GSE20123 | RGS9BP | 388531 | 7 | 5 | 18 | |
| GSE43470 | RGS9BP | 388531 | 5 | 6 | 32 | |
| GSE46452 | RGS9BP | 388531 | 47 | 1 | 11 | |
| GSE47630 | RGS9BP | 388531 | 8 | 6 | 26 | |
| GSE54993 | RGS9BP | 388531 | 17 | 3 | 50 | |
| GSE54994 | RGS9BP | 388531 | 7 | 9 | 37 | |
| GSE60625 | RGS9BP | 388531 | 9 | 0 | 2 | |
| GSE74703 | RGS9BP | 388531 | 5 | 4 | 27 | |
| GSE74704 | RGS9BP | 388531 | 6 | 3 | 11 | |
| TCGA | RGS9BP | 388531 | 18 | 12 | 66 |
Total number of gains: 136; Total number of losses: 55; Total Number of normals: 297.
Somatic mutations of RGS9BP:
Generating mutation plots.
Highly correlated genes for RGS9BP:
Showing top 20/431 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGS9BP | VPS37B | 0.666294 | 4 | 0 | 4 |
| RGS9BP | NDUFC1 | 0.654065 | 3 | 0 | 3 |
| RGS9BP | EXD3 | 0.648702 | 4 | 0 | 4 |
| RGS9BP | SLC10A5 | 0.648551 | 3 | 0 | 3 |
| RGS9BP | RPS16 | 0.647679 | 3 | 0 | 3 |
| RGS9BP | APIP | 0.64195 | 3 | 0 | 3 |
| RGS9BP | TMEM80 | 0.641181 | 5 | 0 | 5 |
| RGS9BP | NR2F6 | 0.640177 | 3 | 0 | 3 |
| RGS9BP | UBL5 | 0.634258 | 3 | 0 | 3 |
| RGS9BP | JPH2 | 0.631371 | 3 | 0 | 3 |
| RGS9BP | YBEY | 0.631369 | 3 | 0 | 3 |
| RGS9BP | ZDHHC3 | 0.629459 | 4 | 0 | 4 |
| RGS9BP | GOT1 | 0.62924 | 3 | 0 | 3 |
| RGS9BP | CLOCK | 0.627119 | 4 | 0 | 4 |
| RGS9BP | FBXO9 | 0.624544 | 4 | 0 | 3 |
| RGS9BP | ARF5 | 0.62437 | 4 | 0 | 3 |
| RGS9BP | CRISP2 | 0.6235 | 3 | 0 | 3 |
| RGS9BP | CCDC12 | 0.622426 | 5 | 0 | 4 |
| RGS9BP | DYM | 0.620753 | 3 | 0 | 3 |
| RGS9BP | KLHDC8A | 0.619542 | 6 | 0 | 4 |
For details and further investigation, click here