| Full name: renalase, FAD dependent amine oxidase | Alias Symbol: FLJ11218|renalase | ||
| Type: protein-coding gene | Cytoband: 10q23.31 | ||
| Entrez ID: 55328 | HGNC ID: HGNC:25641 | Ensembl Gene: ENSG00000184719 | OMIM ID: 609360 |
| Drug and gene relationship at DGIdb | |||
Expression of RNLS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RNLS | 55328 | 223824_at | -0.3750 | 0.2038 | |
| GSE20347 | RNLS | 55328 | 220564_at | 0.0200 | 0.7736 | |
| GSE23400 | RNLS | 55328 | 220564_at | 0.0063 | 0.7708 | |
| GSE26886 | RNLS | 55328 | 223824_at | -1.1296 | 0.0012 | |
| GSE29001 | RNLS | 55328 | 220564_at | 0.0267 | 0.8665 | |
| GSE38129 | RNLS | 55328 | 220564_at | -0.0638 | 0.2441 | |
| GSE45670 | RNLS | 55328 | 223824_at | -0.3879 | 0.1636 | |
| GSE53622 | RNLS | 55328 | 84768 | -1.0225 | 0.0000 | |
| GSE53624 | RNLS | 55328 | 84768 | -0.8091 | 0.0000 | |
| GSE63941 | RNLS | 55328 | 223824_at | -1.7047 | 0.0888 | |
| GSE77861 | RNLS | 55328 | 223824_at | -0.6173 | 0.0081 | |
| GSE97050 | RNLS | 55328 | A_23_P202501 | -0.1738 | 0.5403 | |
| SRP007169 | RNLS | 55328 | RNAseq | -3.3198 | 0.0000 | |
| SRP064894 | RNLS | 55328 | RNAseq | -0.8832 | 0.0003 | |
| SRP133303 | RNLS | 55328 | RNAseq | -0.1272 | 0.2963 | |
| SRP159526 | RNLS | 55328 | RNAseq | -0.4254 | 0.2706 | |
| SRP193095 | RNLS | 55328 | RNAseq | -0.4540 | 0.0259 | |
| SRP219564 | RNLS | 55328 | RNAseq | -0.4573 | 0.2861 | |
| TCGA | RNLS | 55328 | RNAseq | -0.4774 | 0.0302 |
Upregulated datasets: 0; Downregulated datasets: 3.
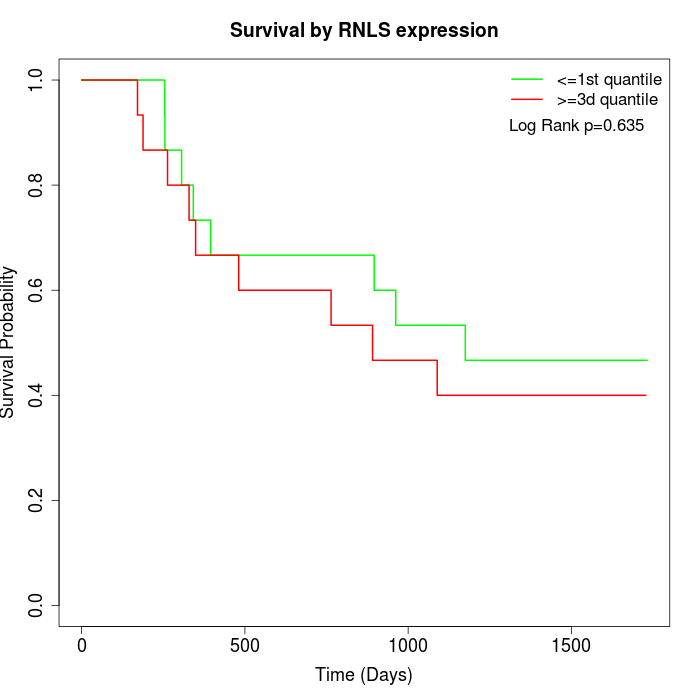
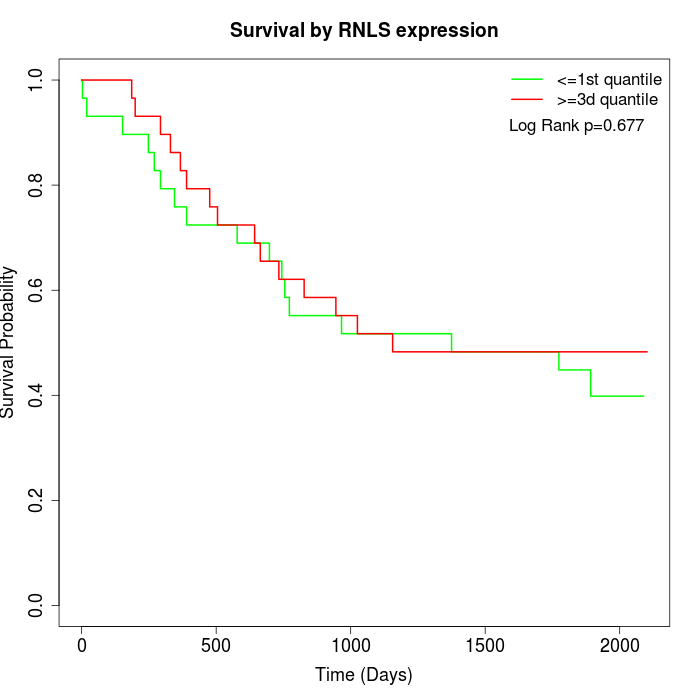
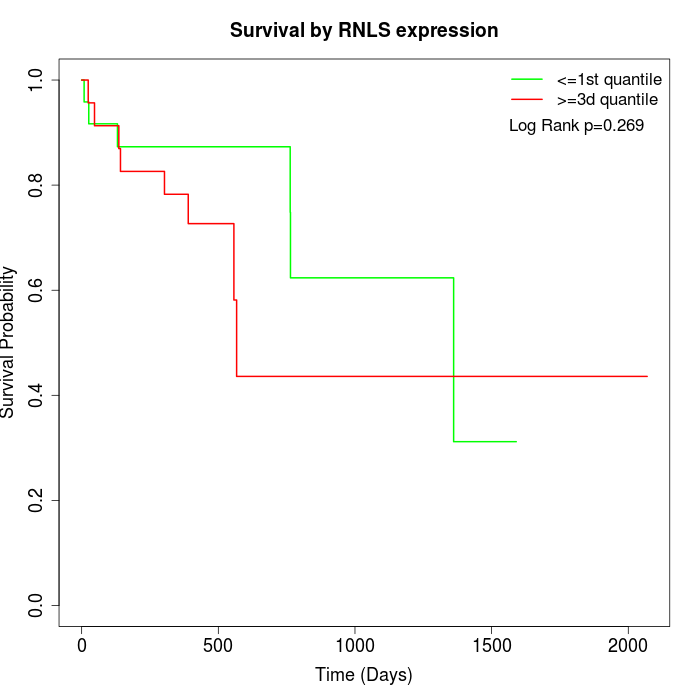
Survival by RNLS expression:
Note: Click image to view full size file.
Copy number change of RNLS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RNLS | 55328 | 2 | 7 | 21 | |
| GSE20123 | RNLS | 55328 | 2 | 6 | 22 | |
| GSE43470 | RNLS | 55328 | 0 | 8 | 35 | |
| GSE46452 | RNLS | 55328 | 0 | 11 | 48 | |
| GSE47630 | RNLS | 55328 | 2 | 14 | 24 | |
| GSE54993 | RNLS | 55328 | 7 | 0 | 63 | |
| GSE54994 | RNLS | 55328 | 1 | 11 | 41 | |
| GSE60625 | RNLS | 55328 | 0 | 0 | 11 | |
| GSE74703 | RNLS | 55328 | 0 | 6 | 30 | |
| GSE74704 | RNLS | 55328 | 1 | 4 | 15 | |
| TCGA | RNLS | 55328 | 7 | 27 | 62 |
Total number of gains: 22; Total number of losses: 94; Total Number of normals: 372.
Somatic mutations of RNLS:
Generating mutation plots.
Highly correlated genes for RNLS:
Showing top 20/952 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RNLS | RHBDD1 | 0.789988 | 3 | 0 | 3 |
| RNLS | PRPF38A | 0.779682 | 3 | 0 | 3 |
| RNLS | KLF4 | 0.778896 | 4 | 0 | 4 |
| RNLS | TMEM126A | 0.778893 | 3 | 0 | 3 |
| RNLS | ESYT2 | 0.778558 | 4 | 0 | 4 |
| RNLS | ANO8 | 0.772232 | 3 | 0 | 3 |
| RNLS | ZFC3H1 | 0.769176 | 4 | 0 | 4 |
| RNLS | DNAH1 | 0.749135 | 3 | 0 | 3 |
| RNLS | PLCD3 | 0.747707 | 3 | 0 | 3 |
| RNLS | FBXO10 | 0.743239 | 3 | 0 | 3 |
| RNLS | INSRR | 0.739488 | 3 | 0 | 3 |
| RNLS | ELP6 | 0.725942 | 3 | 0 | 3 |
| RNLS | P4HTM | 0.72481 | 3 | 0 | 3 |
| RNLS | POLK | 0.716335 | 3 | 0 | 3 |
| RNLS | ZDHHC3 | 0.715791 | 4 | 0 | 3 |
| RNLS | COG5 | 0.714883 | 4 | 0 | 4 |
| RNLS | SHB | 0.714186 | 3 | 0 | 3 |
| RNLS | HEMK1 | 0.713137 | 4 | 0 | 4 |
| RNLS | TSPYL1 | 0.709843 | 4 | 0 | 4 |
| RNLS | EEA1 | 0.706884 | 7 | 0 | 7 |
For details and further investigation, click here