| Full name: Ras related GTP binding D | Alias Symbol: DKFZP761H171|bA11D8.2.1 | ||
| Type: protein-coding gene | Cytoband: 6q15 | ||
| Entrez ID: 58528 | HGNC ID: HGNC:19903 | Ensembl Gene: ENSG00000025039 | OMIM ID: 608268 |
| Drug and gene relationship at DGIdb | |||
RRAGD involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway |
Expression of RRAGD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRAGD | 58528 | 221524_s_at | -2.2030 | 0.0090 | |
| GSE20347 | RRAGD | 58528 | 221524_s_at | -2.4767 | 0.0000 | |
| GSE23400 | RRAGD | 58528 | 221524_s_at | -1.1703 | 0.0000 | |
| GSE26886 | RRAGD | 58528 | 221524_s_at | -3.7932 | 0.0000 | |
| GSE29001 | RRAGD | 58528 | 221524_s_at | -2.5144 | 0.0000 | |
| GSE38129 | RRAGD | 58528 | 221524_s_at | -1.6433 | 0.0000 | |
| GSE45670 | RRAGD | 58528 | 221524_s_at | -1.2896 | 0.0099 | |
| GSE53622 | RRAGD | 58528 | 55744 | -2.1664 | 0.0000 | |
| GSE53624 | RRAGD | 58528 | 55744 | -2.4153 | 0.0000 | |
| GSE63941 | RRAGD | 58528 | 221524_s_at | -0.2523 | 0.8575 | |
| GSE77861 | RRAGD | 58528 | 221524_s_at | -1.9827 | 0.0024 | |
| GSE97050 | RRAGD | 58528 | A_23_P133691 | -0.9634 | 0.1403 | |
| SRP007169 | RRAGD | 58528 | RNAseq | -2.3205 | 0.0025 | |
| SRP008496 | RRAGD | 58528 | RNAseq | -2.5049 | 0.0000 | |
| SRP064894 | RRAGD | 58528 | RNAseq | -1.6892 | 0.0000 | |
| SRP133303 | RRAGD | 58528 | RNAseq | -1.8765 | 0.0000 | |
| SRP159526 | RRAGD | 58528 | RNAseq | -1.6127 | 0.0000 | |
| SRP193095 | RRAGD | 58528 | RNAseq | -2.2186 | 0.0000 | |
| SRP219564 | RRAGD | 58528 | RNAseq | -1.8707 | 0.0000 | |
| TCGA | RRAGD | 58528 | RNAseq | 0.1460 | 0.3687 |
Upregulated datasets: 0; Downregulated datasets: 17.
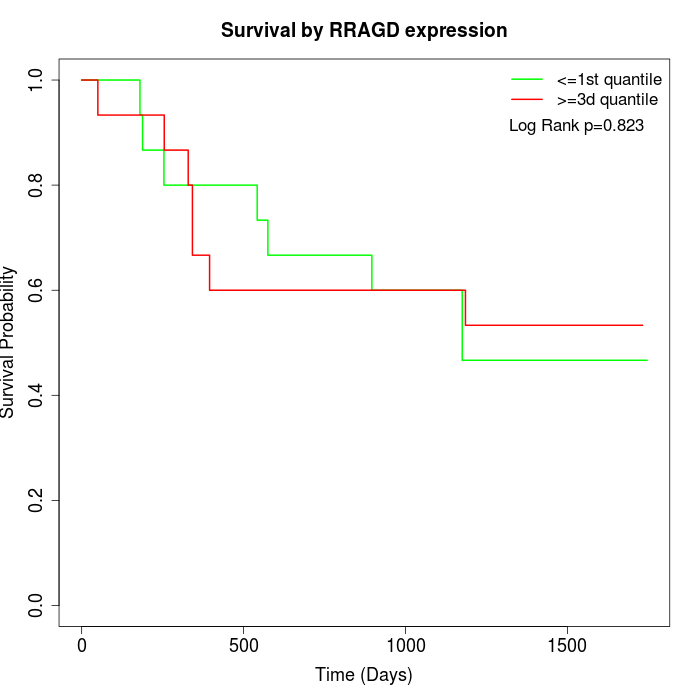
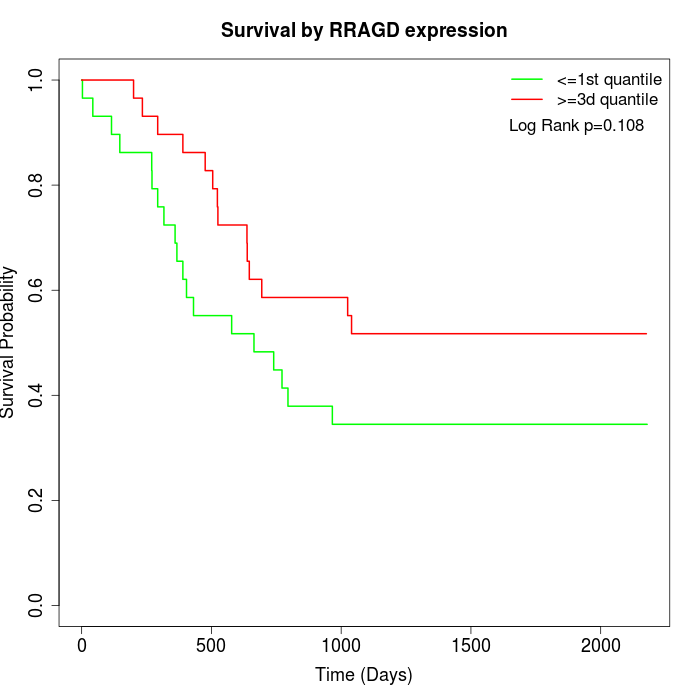
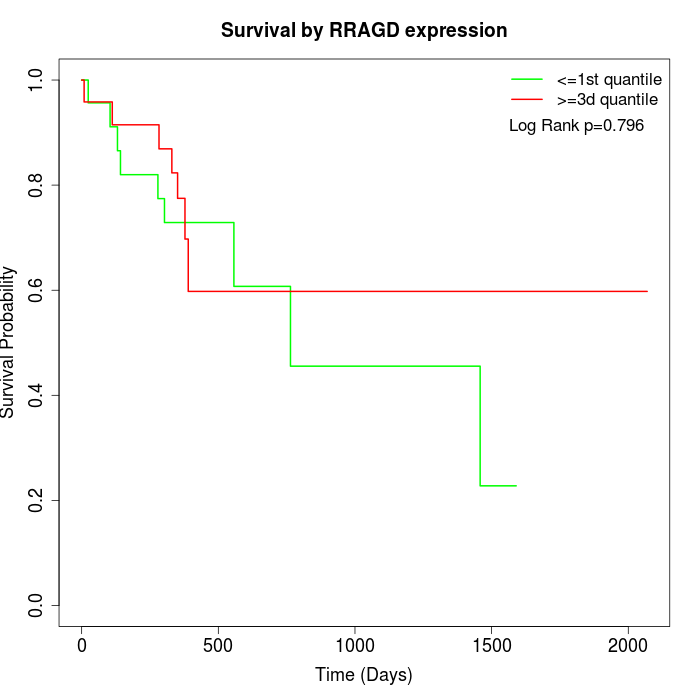
Survival by RRAGD expression:
Note: Click image to view full size file.
Copy number change of RRAGD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RRAGD | 58528 | 1 | 4 | 25 | |
| GSE20123 | RRAGD | 58528 | 1 | 3 | 26 | |
| GSE43470 | RRAGD | 58528 | 3 | 1 | 39 | |
| GSE46452 | RRAGD | 58528 | 2 | 11 | 46 | |
| GSE47630 | RRAGD | 58528 | 8 | 6 | 26 | |
| GSE54993 | RRAGD | 58528 | 3 | 2 | 65 | |
| GSE54994 | RRAGD | 58528 | 8 | 8 | 37 | |
| GSE60625 | RRAGD | 58528 | 0 | 1 | 10 | |
| GSE74703 | RRAGD | 58528 | 3 | 1 | 32 | |
| GSE74704 | RRAGD | 58528 | 0 | 2 | 18 | |
| TCGA | RRAGD | 58528 | 6 | 21 | 69 |
Total number of gains: 35; Total number of losses: 60; Total Number of normals: 393.
Somatic mutations of RRAGD:
Generating mutation plots.
Highly correlated genes for RRAGD:
Showing top 20/1878 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRAGD | RMND5B | 0.848912 | 11 | 0 | 11 |
| RRAGD | CTTNBP2 | 0.843906 | 6 | 0 | 6 |
| RRAGD | KAT2B | 0.842731 | 11 | 0 | 11 |
| RRAGD | SH3BGRL2 | 0.841465 | 7 | 0 | 7 |
| RRAGD | PRSS27 | 0.838751 | 6 | 0 | 6 |
| RRAGD | VSIG10L | 0.837978 | 6 | 0 | 6 |
| RRAGD | SASH1 | 0.834131 | 11 | 0 | 11 |
| RRAGD | TMPRSS11B | 0.829272 | 6 | 0 | 6 |
| RRAGD | SLURP1 | 0.827614 | 10 | 0 | 10 |
| RRAGD | CXCR2 | 0.827522 | 11 | 0 | 11 |
| RRAGD | CASC3 | 0.82676 | 3 | 0 | 3 |
| RRAGD | SNORA68 | 0.824755 | 4 | 0 | 4 |
| RRAGD | FCHO2 | 0.823341 | 7 | 0 | 7 |
| RRAGD | SLC16A6 | 0.82154 | 11 | 0 | 11 |
| RRAGD | SCNN1B | 0.821534 | 10 | 0 | 10 |
| RRAGD | CA13 | 0.820554 | 3 | 0 | 3 |
| RRAGD | SIM2 | 0.81881 | 11 | 0 | 11 |
| RRAGD | SPINK7 | 0.818016 | 6 | 0 | 6 |
| RRAGD | CAPN14 | 0.815632 | 6 | 0 | 6 |
| RRAGD | SFTA2 | 0.813129 | 4 | 0 | 4 |
For details and further investigation, click here