| Full name: ryanodine receptor 1 | Alias Symbol: RYR|PPP1R137 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 6261 | HGNC ID: HGNC:10483 | Ensembl Gene: ENSG00000196218 | OMIM ID: 180901 |
| Drug and gene relationship at DGIdb | |||
RYR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04713 | Circadian entrainment | |
| hsa04730 | Long-term depression | |
| hsa04921 | Oxytocin signaling pathway |
Expression of RYR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RYR1 | 6261 | 205485_at | 0.7129 | 0.3330 | |
| GSE20347 | RYR1 | 6261 | 205485_at | 0.3944 | 0.3221 | |
| GSE23400 | RYR1 | 6261 | 205485_at | 0.6986 | 0.0000 | |
| GSE26886 | RYR1 | 6261 | 205485_at | 0.1254 | 0.7596 | |
| GSE29001 | RYR1 | 6261 | 205485_at | 0.8621 | 0.0338 | |
| GSE38129 | RYR1 | 6261 | 205485_at | 1.0699 | 0.0012 | |
| GSE45670 | RYR1 | 6261 | 205485_at | 0.5715 | 0.0065 | |
| GSE53622 | RYR1 | 6261 | 1428 | -2.1200 | 0.0000 | |
| GSE53624 | RYR1 | 6261 | 1428 | -2.1980 | 0.0000 | |
| GSE63941 | RYR1 | 6261 | 205485_at | 1.9114 | 0.1657 | |
| GSE77861 | RYR1 | 6261 | 205485_at | 0.6076 | 0.3743 | |
| GSE97050 | RYR1 | 6261 | A_23_P78867 | -0.3294 | 0.6968 | |
| SRP007169 | RYR1 | 6261 | RNAseq | 1.3460 | 0.0857 | |
| SRP064894 | RYR1 | 6261 | RNAseq | 1.1990 | 0.0014 | |
| SRP133303 | RYR1 | 6261 | RNAseq | 0.0778 | 0.8387 | |
| SRP159526 | RYR1 | 6261 | RNAseq | 0.9904 | 0.0002 | |
| SRP193095 | RYR1 | 6261 | RNAseq | -0.0945 | 0.7591 | |
| SRP219564 | RYR1 | 6261 | RNAseq | 0.3976 | 0.5523 | |
| TCGA | RYR1 | 6261 | RNAseq | 1.1457 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 2.
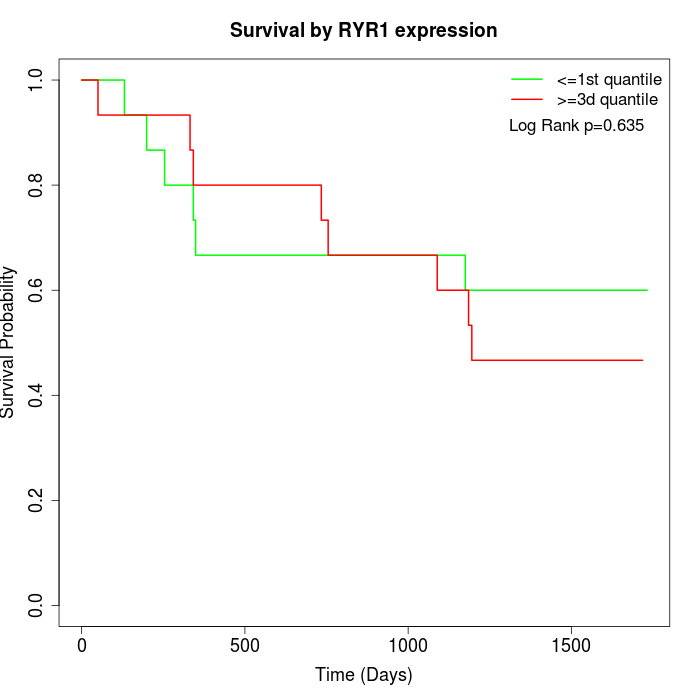
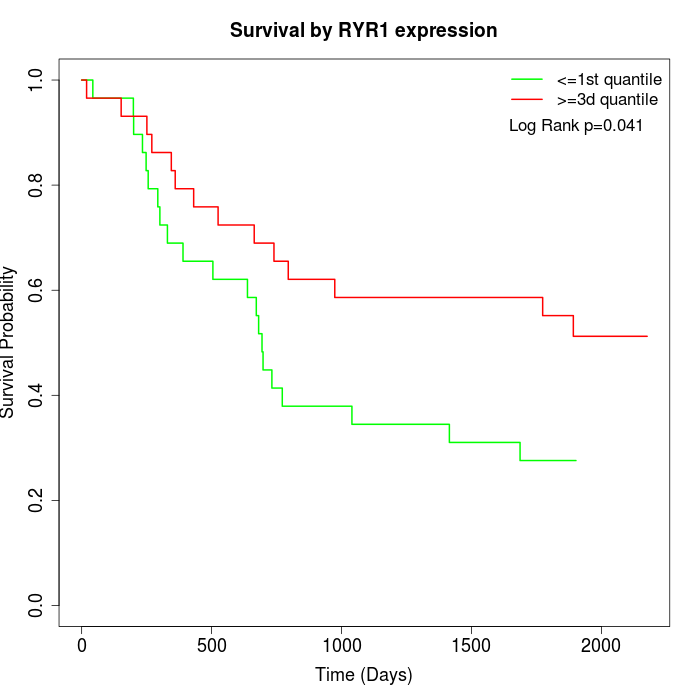
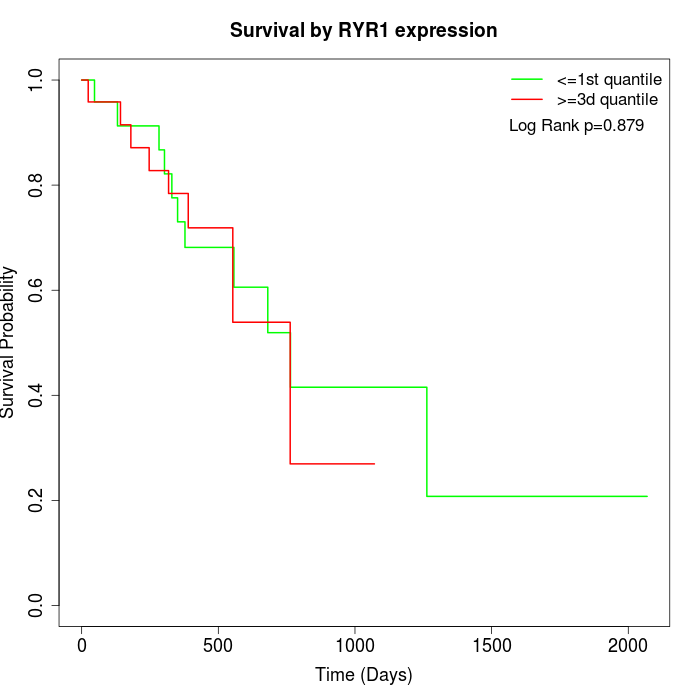
Survival by RYR1 expression:
Note: Click image to view full size file.
Copy number change of RYR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RYR1 | 6261 | 7 | 4 | 19 | |
| GSE20123 | RYR1 | 6261 | 7 | 3 | 20 | |
| GSE43470 | RYR1 | 6261 | 4 | 10 | 29 | |
| GSE46452 | RYR1 | 6261 | 47 | 2 | 10 | |
| GSE47630 | RYR1 | 6261 | 9 | 5 | 26 | |
| GSE54993 | RYR1 | 6261 | 17 | 3 | 50 | |
| GSE54994 | RYR1 | 6261 | 6 | 9 | 38 | |
| GSE60625 | RYR1 | 6261 | 9 | 0 | 2 | |
| GSE74703 | RYR1 | 6261 | 4 | 6 | 26 | |
| GSE74704 | RYR1 | 6261 | 7 | 1 | 12 | |
| TCGA | RYR1 | 6261 | 16 | 15 | 65 |
Total number of gains: 133; Total number of losses: 58; Total Number of normals: 297.
Somatic mutations of RYR1:
Generating mutation plots.
Highly correlated genes for RYR1:
Showing top 20/77 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RYR1 | SLC4A3 | 0.784285 | 3 | 0 | 3 |
| RYR1 | KRTAP4-1 | 0.692716 | 5 | 0 | 5 |
| RYR1 | SFXN3 | 0.666556 | 4 | 0 | 4 |
| RYR1 | RRP1 | 0.647719 | 4 | 0 | 3 |
| RYR1 | ERICH4 | 0.642982 | 3 | 0 | 3 |
| RYR1 | CTBP1 | 0.630807 | 4 | 0 | 4 |
| RYR1 | CEACAM19 | 0.623542 | 3 | 0 | 3 |
| RYR1 | ZNF114 | 0.620402 | 4 | 0 | 3 |
| RYR1 | ALKBH6 | 0.617169 | 4 | 0 | 3 |
| RYR1 | TNFRSF1A | 0.612462 | 3 | 0 | 3 |
| RYR1 | EP400 | 0.610753 | 3 | 0 | 3 |
| RYR1 | TFAP2E | 0.61037 | 4 | 0 | 3 |
| RYR1 | USP14 | 0.602719 | 5 | 0 | 3 |
| RYR1 | PHF12 | 0.601748 | 5 | 0 | 3 |
| RYR1 | ESYT3 | 0.601449 | 5 | 0 | 3 |
| RYR1 | MDFI | 0.595883 | 4 | 0 | 3 |
| RYR1 | KCTD15 | 0.595554 | 4 | 0 | 4 |
| RYR1 | FOXRED1 | 0.593218 | 4 | 0 | 3 |
| RYR1 | UVRAG | 0.592987 | 5 | 0 | 3 |
| RYR1 | BNC1 | 0.581328 | 4 | 0 | 3 |
For details and further investigation, click here