| Full name: sterile alpha motif domain containing 13 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p31.1 | ||
| Entrez ID: 148418 | HGNC ID: HGNC:24582 | Ensembl Gene: ENSG00000203943 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SAMD13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SAMD13 | 148418 | 229402_at | -0.1772 | 0.7954 | |
| GSE26886 | SAMD13 | 148418 | 229402_at | -0.2422 | 0.1947 | |
| GSE45670 | SAMD13 | 148418 | 229402_at | -0.3863 | 0.0501 | |
| GSE53622 | SAMD13 | 148418 | 26895 | -0.5169 | 0.0002 | |
| GSE53624 | SAMD13 | 148418 | 26895 | -0.7685 | 0.0000 | |
| GSE63941 | SAMD13 | 148418 | 229402_at | 0.5471 | 0.5347 | |
| GSE77861 | SAMD13 | 148418 | 229402_at | -0.1533 | 0.2655 | |
| GSE97050 | SAMD13 | 148418 | A_33_P3266429 | -0.0073 | 0.9790 | |
| SRP133303 | SAMD13 | 148418 | RNAseq | -0.6825 | 0.0333 | |
| SRP159526 | SAMD13 | 148418 | RNAseq | -1.4615 | 0.0018 | |
| TCGA | SAMD13 | 148418 | RNAseq | -1.7840 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
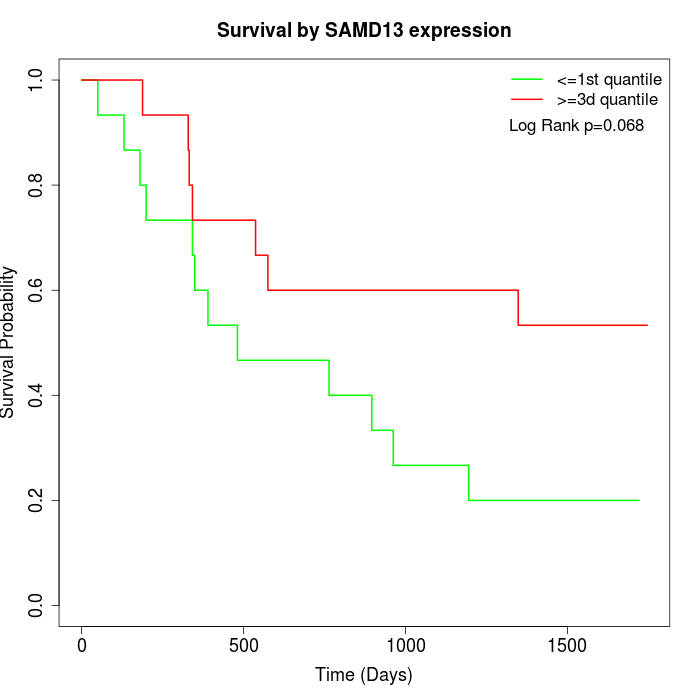
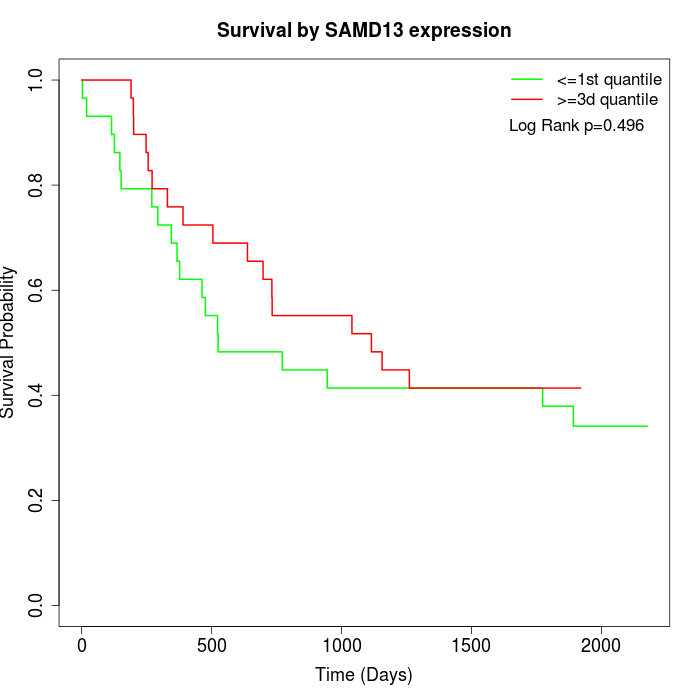
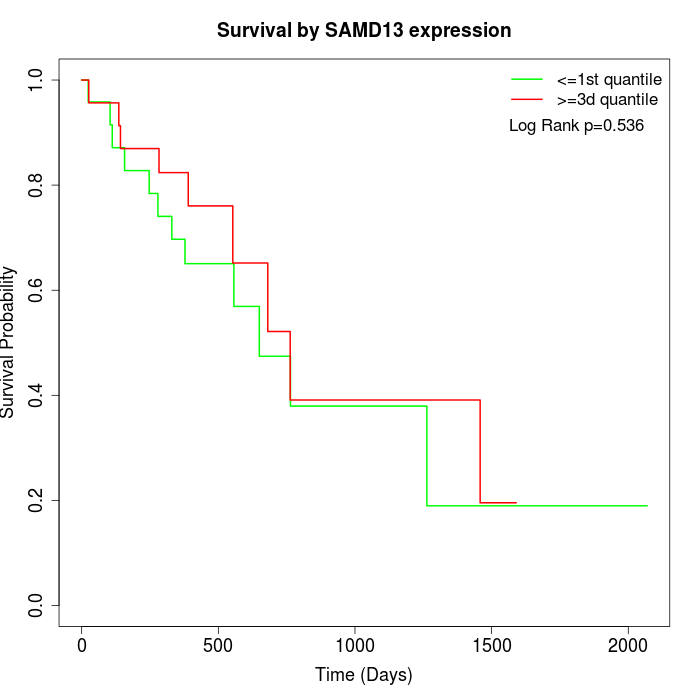
Survival by SAMD13 expression:
Note: Click image to view full size file.
Copy number change of SAMD13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SAMD13 | 148418 | 0 | 8 | 22 | |
| GSE20123 | SAMD13 | 148418 | 0 | 8 | 22 | |
| GSE43470 | SAMD13 | 148418 | 2 | 2 | 39 | |
| GSE46452 | SAMD13 | 148418 | 0 | 1 | 58 | |
| GSE47630 | SAMD13 | 148418 | 8 | 5 | 27 | |
| GSE54993 | SAMD13 | 148418 | 0 | 0 | 70 | |
| GSE54994 | SAMD13 | 148418 | 6 | 3 | 44 | |
| GSE60625 | SAMD13 | 148418 | 0 | 0 | 11 | |
| GSE74703 | SAMD13 | 148418 | 1 | 2 | 33 | |
| GSE74704 | SAMD13 | 148418 | 0 | 5 | 15 | |
| TCGA | SAMD13 | 148418 | 8 | 23 | 65 |
Total number of gains: 25; Total number of losses: 57; Total Number of normals: 406.
Somatic mutations of SAMD13:
Generating mutation plots.
Highly correlated genes for SAMD13:
Showing top 20/41 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SAMD13 | MED11 | 0.719027 | 3 | 0 | 3 |
| SAMD13 | C7orf26 | 0.717291 | 3 | 0 | 3 |
| SAMD13 | KIAA1671 | 0.676343 | 3 | 0 | 3 |
| SAMD13 | METTL18 | 0.640604 | 3 | 0 | 3 |
| SAMD13 | FA2H | 0.61243 | 3 | 0 | 3 |
| SAMD13 | LARP4B | 0.607714 | 4 | 0 | 3 |
| SAMD13 | MEF2C | 0.605411 | 3 | 0 | 3 |
| SAMD13 | ACTR1B | 0.591857 | 3 | 0 | 3 |
| SAMD13 | PPOX | 0.590475 | 5 | 0 | 3 |
| SAMD13 | CLDN8 | 0.589313 | 4 | 0 | 3 |
| SAMD13 | C15orf65 | 0.586575 | 3 | 0 | 3 |
| SAMD13 | BCL2L15 | 0.586436 | 4 | 0 | 3 |
| SAMD13 | TM9SF2 | 0.585632 | 3 | 0 | 3 |
| SAMD13 | EEF1A1 | 0.580364 | 3 | 0 | 3 |
| SAMD13 | BTBD9 | 0.570753 | 5 | 0 | 3 |
| SAMD13 | ZADH2 | 0.567158 | 4 | 0 | 3 |
| SAMD13 | TSPAN12 | 0.567123 | 4 | 0 | 3 |
| SAMD13 | ZMYND12 | 0.566004 | 6 | 0 | 5 |
| SAMD13 | MANEAL | 0.564821 | 4 | 0 | 3 |
| SAMD13 | C9orf152 | 0.563877 | 3 | 0 | 3 |
For details and further investigation, click here