| Full name: zinc finger MYND-type containing 12 | Alias Symbol: DKFZp434N2435 | ||
| Type: protein-coding gene | Cytoband: 1p34.2 | ||
| Entrez ID: 84217 | HGNC ID: HGNC:21192 | Ensembl Gene: ENSG00000066185 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ZMYND12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZMYND12 | 84217 | 223636_at | -0.1840 | 0.6053 | |
| GSE26886 | ZMYND12 | 84217 | 223636_at | 0.0265 | 0.8888 | |
| GSE45670 | ZMYND12 | 84217 | 223636_at | -0.1315 | 0.4173 | |
| GSE53622 | ZMYND12 | 84217 | 44934 | -0.2626 | 0.0181 | |
| GSE53624 | ZMYND12 | 84217 | 44934 | -0.3886 | 0.0014 | |
| GSE63941 | ZMYND12 | 84217 | 223636_at | -0.2260 | 0.7267 | |
| GSE77861 | ZMYND12 | 84217 | 223636_at | -0.0499 | 0.5574 | |
| GSE97050 | ZMYND12 | 84217 | A_23_P160567 | 0.2892 | 0.1955 | |
| SRP159526 | ZMYND12 | 84217 | RNAseq | -0.2961 | 0.6176 | |
| TCGA | ZMYND12 | 84217 | RNAseq | -1.3318 | 0.0009 |
Upregulated datasets: 0; Downregulated datasets: 1.
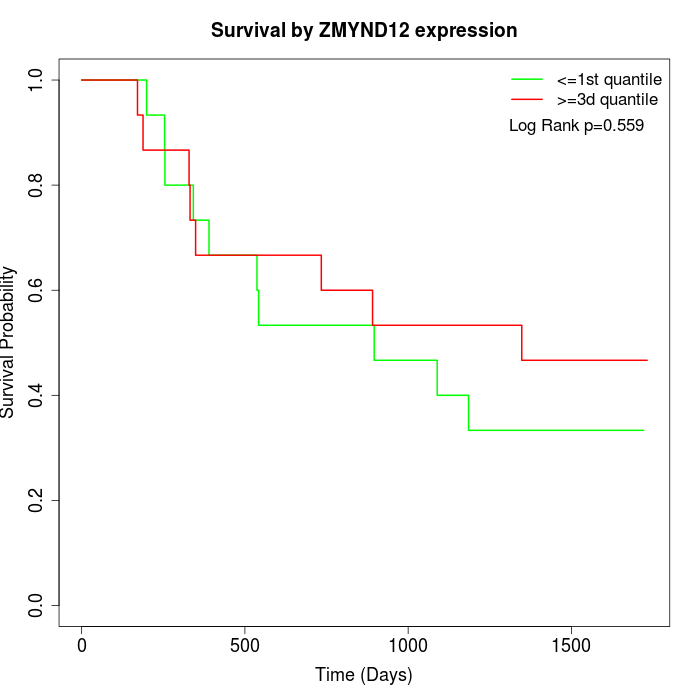
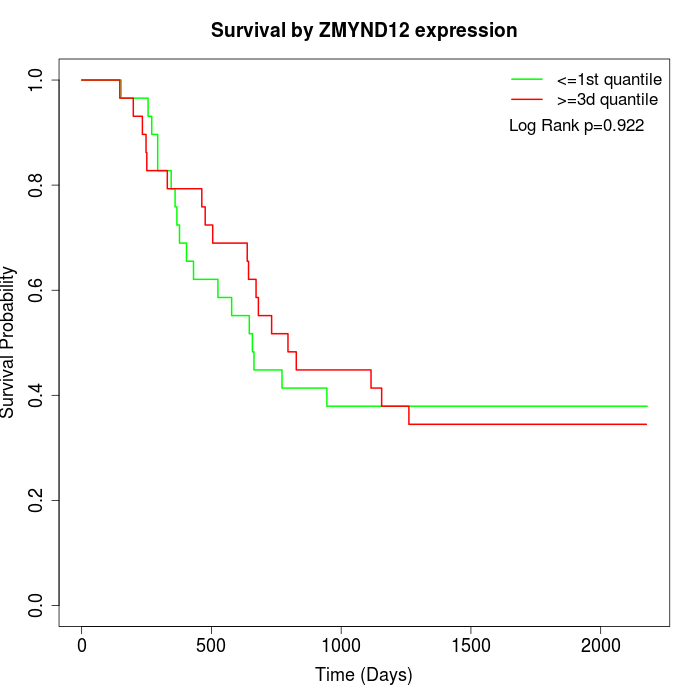
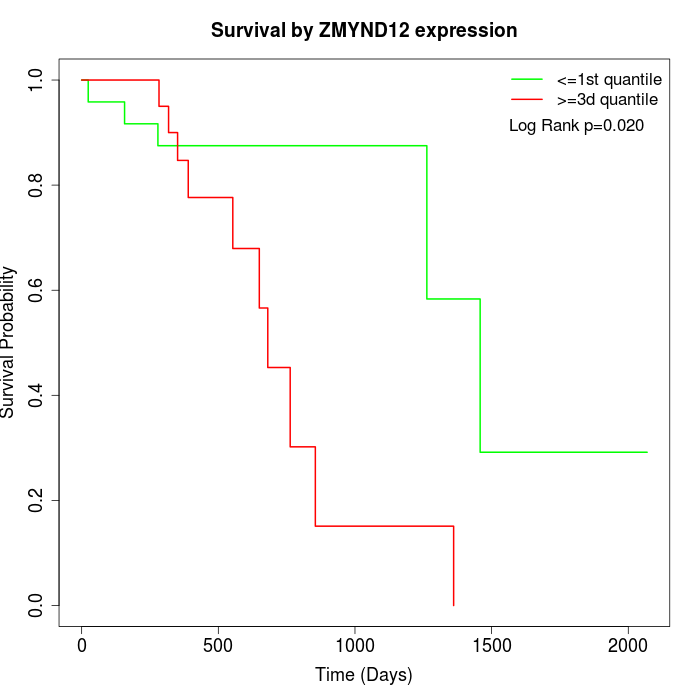
Survival by ZMYND12 expression:
Note: Click image to view full size file.
Copy number change of ZMYND12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZMYND12 | 84217 | 4 | 3 | 23 | |
| GSE20123 | ZMYND12 | 84217 | 3 | 2 | 25 | |
| GSE43470 | ZMYND12 | 84217 | 7 | 2 | 34 | |
| GSE46452 | ZMYND12 | 84217 | 3 | 1 | 55 | |
| GSE47630 | ZMYND12 | 84217 | 9 | 2 | 29 | |
| GSE54993 | ZMYND12 | 84217 | 0 | 2 | 68 | |
| GSE54994 | ZMYND12 | 84217 | 13 | 2 | 38 | |
| GSE60625 | ZMYND12 | 84217 | 0 | 0 | 11 | |
| GSE74703 | ZMYND12 | 84217 | 6 | 1 | 29 | |
| GSE74704 | ZMYND12 | 84217 | 1 | 0 | 19 | |
| TCGA | ZMYND12 | 84217 | 13 | 16 | 67 |
Total number of gains: 59; Total number of losses: 31; Total Number of normals: 398.
Somatic mutations of ZMYND12:
Generating mutation plots.
Highly correlated genes for ZMYND12:
Showing top 20/22 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZMYND12 | NRIP2 | 0.687747 | 3 | 0 | 3 |
| ZMYND12 | SKAP1 | 0.670916 | 3 | 0 | 3 |
| ZMYND12 | NKX2-8 | 0.666683 | 3 | 0 | 3 |
| ZMYND12 | RIPK4 | 0.654034 | 3 | 0 | 3 |
| ZMYND12 | LIMD1 | 0.653066 | 3 | 0 | 3 |
| ZMYND12 | TAMM41 | 0.626601 | 4 | 0 | 3 |
| ZMYND12 | LENG9 | 0.623228 | 3 | 0 | 3 |
| ZMYND12 | UBN2 | 0.617437 | 3 | 0 | 3 |
| ZMYND12 | FUZ | 0.597906 | 4 | 0 | 3 |
| ZMYND12 | IFI27L1 | 0.587366 | 3 | 0 | 3 |
| ZMYND12 | PLD4 | 0.579274 | 4 | 0 | 3 |
| ZMYND12 | ZBTB3 | 0.576432 | 5 | 0 | 4 |
| ZMYND12 | SAMD13 | 0.566004 | 6 | 0 | 5 |
| ZMYND12 | FHIT | 0.558532 | 4 | 0 | 3 |
| ZMYND12 | ADAMTSL3 | 0.554121 | 4 | 0 | 3 |
| ZMYND12 | SIGLEC8 | 0.553935 | 3 | 0 | 3 |
| ZMYND12 | VIPR2 | 0.54513 | 5 | 0 | 3 |
| ZMYND12 | ICA1 | 0.536548 | 6 | 0 | 3 |
| ZMYND12 | ABAT | 0.531159 | 5 | 0 | 3 |
| ZMYND12 | CCDC89 | 0.530664 | 5 | 0 | 3 |
For details and further investigation, click here