| Full name: DNA damage regulated autophagy modulator 1 | Alias Symbol: FLJ11259|DRAM | ||
| Type: protein-coding gene | Cytoband: 12q23.2 | ||
| Entrez ID: 55332 | HGNC ID: HGNC:25645 | Ensembl Gene: ENSG00000136048 | OMIM ID: 610776 |
| Drug and gene relationship at DGIdb | |||
Expression of DRAM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DRAM1 | 55332 | 218627_at | 0.2297 | 0.5791 | |
| GSE20347 | DRAM1 | 55332 | 218627_at | 0.9593 | 0.0000 | |
| GSE23400 | DRAM1 | 55332 | 218627_at | 0.7384 | 0.0000 | |
| GSE26886 | DRAM1 | 55332 | 218627_at | 1.3270 | 0.0012 | |
| GSE29001 | DRAM1 | 55332 | 218627_at | 0.8807 | 0.0057 | |
| GSE38129 | DRAM1 | 55332 | 218627_at | 0.7575 | 0.0009 | |
| GSE45670 | DRAM1 | 55332 | 218627_at | 0.0115 | 0.9792 | |
| GSE53622 | DRAM1 | 55332 | 69066 | 1.0669 | 0.0000 | |
| GSE53624 | DRAM1 | 55332 | 69066 | 0.9466 | 0.0000 | |
| GSE63941 | DRAM1 | 55332 | 218627_at | -3.6463 | 0.0008 | |
| GSE77861 | DRAM1 | 55332 | 218627_at | 0.4734 | 0.1200 | |
| GSE97050 | DRAM1 | 55332 | A_23_P99163 | 0.6034 | 0.1592 | |
| SRP007169 | DRAM1 | 55332 | RNAseq | 1.9514 | 0.0000 | |
| SRP008496 | DRAM1 | 55332 | RNAseq | 2.1171 | 0.0000 | |
| SRP064894 | DRAM1 | 55332 | RNAseq | 1.2916 | 0.0000 | |
| SRP133303 | DRAM1 | 55332 | RNAseq | 0.9169 | 0.0000 | |
| SRP159526 | DRAM1 | 55332 | RNAseq | 0.8183 | 0.0003 | |
| SRP193095 | DRAM1 | 55332 | RNAseq | 0.8829 | 0.0000 | |
| SRP219564 | DRAM1 | 55332 | RNAseq | 0.9411 | 0.0541 | |
| TCGA | DRAM1 | 55332 | RNAseq | -0.1489 | 0.0891 |
Upregulated datasets: 5; Downregulated datasets: 1.
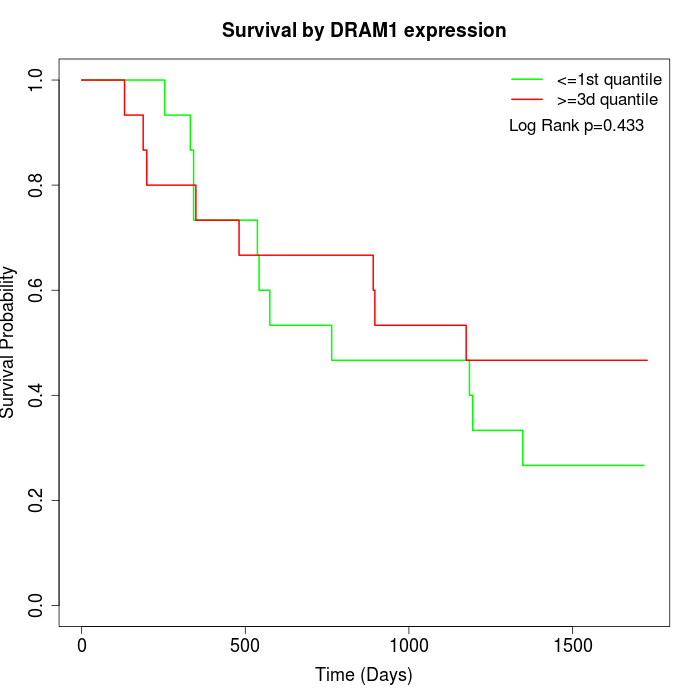
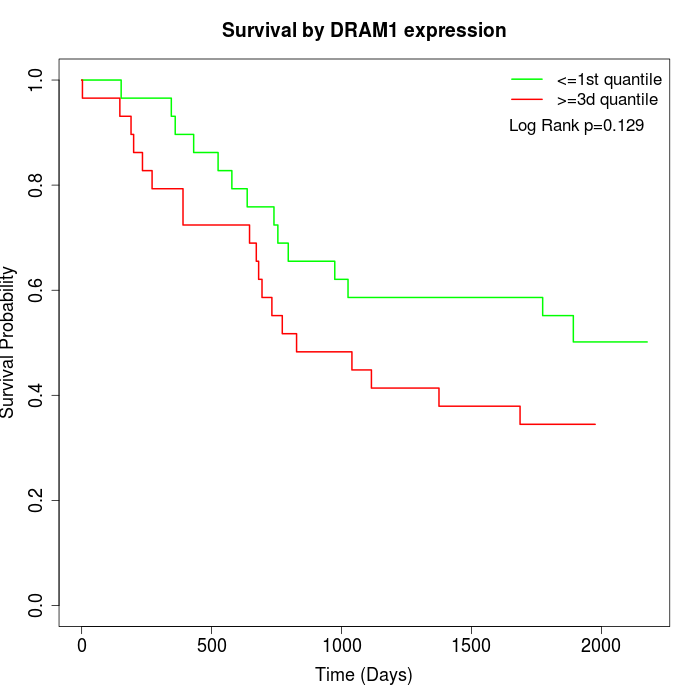
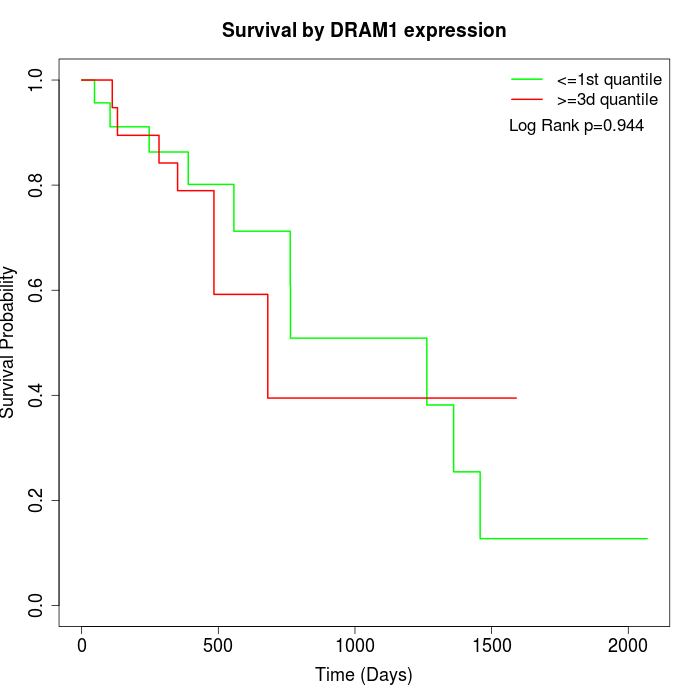
Survival by DRAM1 expression:
Note: Click image to view full size file.
Copy number change of DRAM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DRAM1 | 55332 | 3 | 2 | 25 | |
| GSE20123 | DRAM1 | 55332 | 3 | 2 | 25 | |
| GSE43470 | DRAM1 | 55332 | 2 | 0 | 41 | |
| GSE46452 | DRAM1 | 55332 | 9 | 1 | 49 | |
| GSE47630 | DRAM1 | 55332 | 9 | 1 | 30 | |
| GSE54993 | DRAM1 | 55332 | 0 | 5 | 65 | |
| GSE54994 | DRAM1 | 55332 | 4 | 2 | 47 | |
| GSE60625 | DRAM1 | 55332 | 0 | 0 | 11 | |
| GSE74703 | DRAM1 | 55332 | 2 | 0 | 34 | |
| GSE74704 | DRAM1 | 55332 | 1 | 2 | 17 | |
| TCGA | DRAM1 | 55332 | 18 | 10 | 68 |
Total number of gains: 51; Total number of losses: 25; Total Number of normals: 412.
Somatic mutations of DRAM1:
Generating mutation plots.
Highly correlated genes for DRAM1:
Showing top 20/1469 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DRAM1 | FCGR3A | 0.828196 | 3 | 0 | 3 |
| DRAM1 | LTBR | 0.794424 | 3 | 0 | 3 |
| DRAM1 | LRRC8C | 0.784505 | 3 | 0 | 3 |
| DRAM1 | PDIA5 | 0.765701 | 3 | 0 | 3 |
| DRAM1 | CERKL | 0.747917 | 3 | 0 | 3 |
| DRAM1 | C1S | 0.745673 | 13 | 0 | 13 |
| DRAM1 | SH2B3 | 0.736844 | 13 | 0 | 12 |
| DRAM1 | ZNF469 | 0.733942 | 5 | 0 | 5 |
| DRAM1 | TIMP1 | 0.731929 | 13 | 0 | 12 |
| DRAM1 | RAB34 | 0.728177 | 3 | 0 | 3 |
| DRAM1 | SPON2 | 0.723121 | 3 | 0 | 3 |
| DRAM1 | MTHFD1L | 0.72271 | 4 | 0 | 4 |
| DRAM1 | DACT1 | 0.721905 | 11 | 0 | 11 |
| DRAM1 | FCER1G | 0.721362 | 11 | 0 | 11 |
| DRAM1 | C3AR1 | 0.717138 | 10 | 0 | 10 |
| DRAM1 | LHFPL2 | 0.71682 | 13 | 0 | 11 |
| DRAM1 | HYLS1 | 0.712685 | 3 | 0 | 3 |
| DRAM1 | TNFRSF19 | 0.711675 | 3 | 0 | 3 |
| DRAM1 | TNS3 | 0.710055 | 13 | 0 | 13 |
| DRAM1 | IL2RA | 0.709647 | 5 | 0 | 4 |
For details and further investigation, click here