| Full name: signal regulatory protein alpha | Alias Symbol: SHPS1|SIRP|MYD-1|BIT|P84|SHPS-1|SIRPalpha|CD172a|SIRPalpha2|MFR|SIRP-ALPHA-1 | ||
| Type: protein-coding gene | Cytoband: 20p13 | ||
| Entrez ID: 140885 | HGNC ID: HGNC:9662 | Ensembl Gene: ENSG00000198053 | OMIM ID: 602461 |
| Drug and gene relationship at DGIdb | |||
SIRPA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04380 | Osteoclast differentiation |
Expression of SIRPA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SIRPA | 140885 | 202897_at | 0.7265 | 0.4288 | |
| GSE20347 | SIRPA | 140885 | 202897_at | 0.6609 | 0.0019 | |
| GSE23400 | SIRPA | 140885 | 202896_s_at | 0.3311 | 0.0000 | |
| GSE26886 | SIRPA | 140885 | 202897_at | 0.7703 | 0.0259 | |
| GSE29001 | SIRPA | 140885 | 202897_at | 0.9221 | 0.0102 | |
| GSE38129 | SIRPA | 140885 | 202897_at | 0.6733 | 0.0004 | |
| GSE45670 | SIRPA | 140885 | 202897_at | 0.7909 | 0.0030 | |
| GSE53622 | SIRPA | 140885 | 48773 | 0.7581 | 0.0000 | |
| GSE53624 | SIRPA | 140885 | 48773 | 0.6553 | 0.0000 | |
| GSE63941 | SIRPA | 140885 | 202897_at | -3.8811 | 0.0003 | |
| GSE77861 | SIRPA | 140885 | 202897_at | 0.3262 | 0.5162 | |
| GSE97050 | SIRPA | 140885 | A_23_P210708 | 0.6067 | 0.1506 | |
| SRP007169 | SIRPA | 140885 | RNAseq | 1.6845 | 0.0104 | |
| SRP008496 | SIRPA | 140885 | RNAseq | 1.4491 | 0.0010 | |
| SRP064894 | SIRPA | 140885 | RNAseq | 1.4237 | 0.0000 | |
| SRP133303 | SIRPA | 140885 | RNAseq | 1.1847 | 0.0000 | |
| SRP159526 | SIRPA | 140885 | RNAseq | 0.9759 | 0.0077 | |
| SRP193095 | SIRPA | 140885 | RNAseq | 1.2137 | 0.0000 | |
| SRP219564 | SIRPA | 140885 | RNAseq | 1.4363 | 0.0001 | |
| TCGA | SIRPA | 140885 | RNAseq | 0.3045 | 0.0003 |
Upregulated datasets: 6; Downregulated datasets: 1.
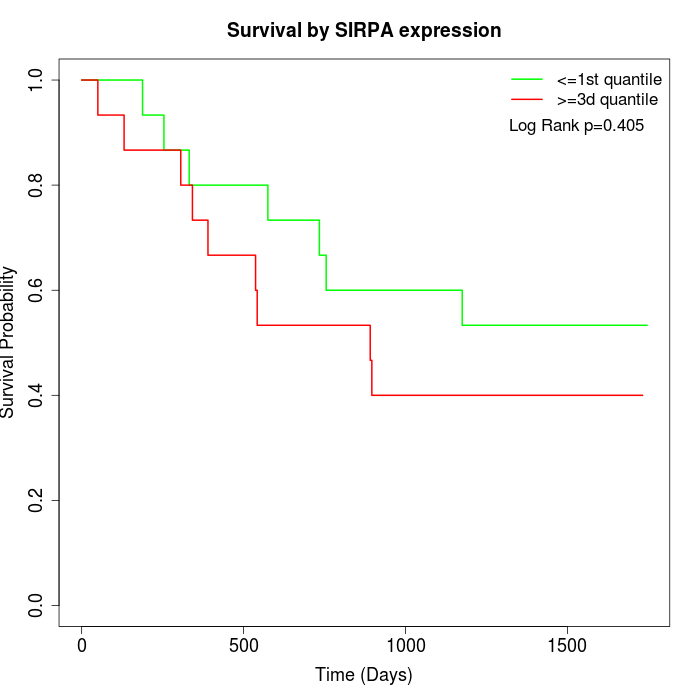
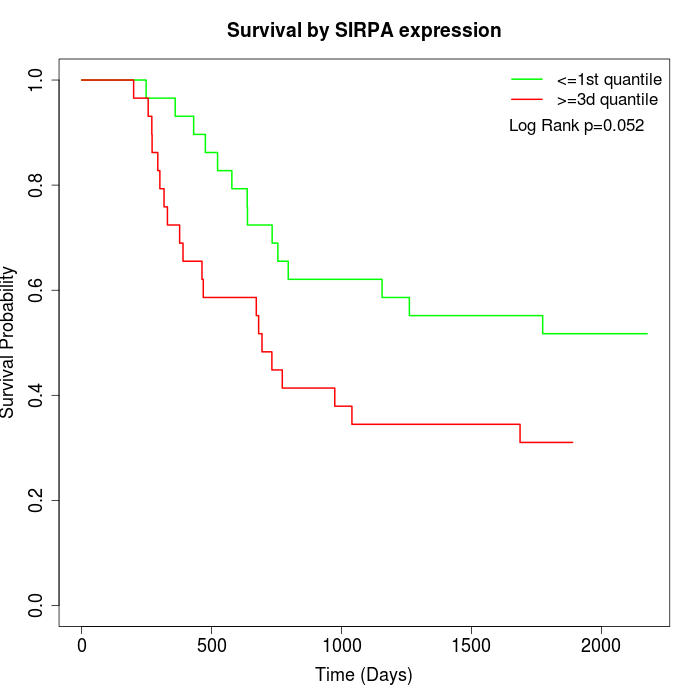
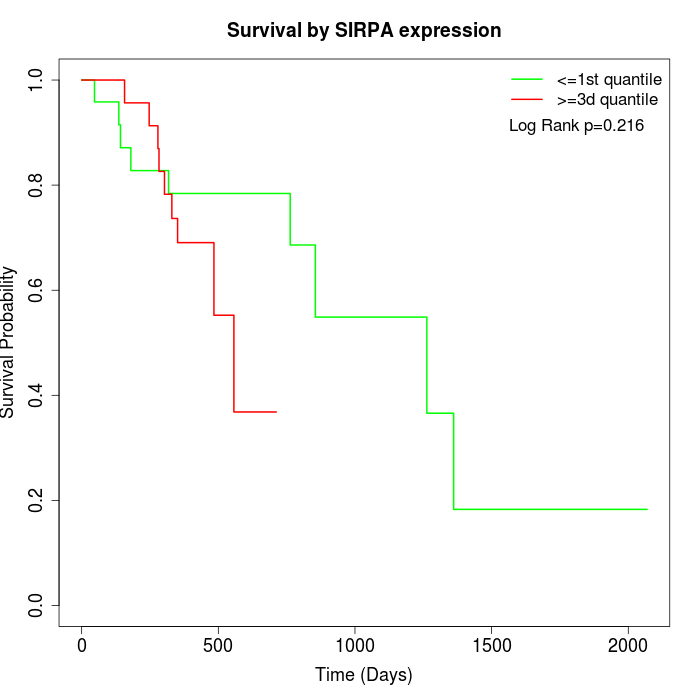
Survival by SIRPA expression:
Note: Click image to view full size file.
Copy number change of SIRPA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SIRPA | 140885 | 12 | 3 | 15 | |
| GSE20123 | SIRPA | 140885 | 13 | 3 | 14 | |
| GSE43470 | SIRPA | 140885 | 11 | 1 | 31 | |
| GSE46452 | SIRPA | 140885 | 26 | 1 | 32 | |
| GSE47630 | SIRPA | 140885 | 18 | 4 | 18 | |
| GSE54993 | SIRPA | 140885 | 2 | 14 | 54 | |
| GSE54994 | SIRPA | 140885 | 24 | 2 | 27 | |
| GSE60625 | SIRPA | 140885 | 0 | 0 | 11 | |
| GSE74703 | SIRPA | 140885 | 10 | 1 | 25 | |
| GSE74704 | SIRPA | 140885 | 8 | 1 | 11 | |
| TCGA | SIRPA | 140885 | 40 | 11 | 45 |
Total number of gains: 164; Total number of losses: 41; Total Number of normals: 283.
Somatic mutations of SIRPA:
Generating mutation plots.
Highly correlated genes for SIRPA:
Showing top 20/294 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SIRPA | NPW | 0.755653 | 3 | 0 | 3 |
| SIRPA | MYO1C | 0.73359 | 3 | 0 | 3 |
| SIRPA | GNRH1 | 0.7014 | 3 | 0 | 3 |
| SIRPA | LOX | 0.685649 | 8 | 0 | 8 |
| SIRPA | IGSF21 | 0.681956 | 3 | 0 | 3 |
| SIRPA | LAYN | 0.680434 | 5 | 0 | 3 |
| SIRPA | ADAMTS12 | 0.673592 | 6 | 0 | 6 |
| SIRPA | SDK1 | 0.67167 | 4 | 0 | 4 |
| SIRPA | RAD51D | 0.669257 | 4 | 0 | 3 |
| SIRPA | OPN3 | 0.666519 | 3 | 0 | 3 |
| SIRPA | PMEPA1 | 0.666216 | 11 | 0 | 9 |
| SIRPA | FABP6 | 0.66358 | 4 | 0 | 3 |
| SIRPA | CHSY3 | 0.659459 | 7 | 0 | 6 |
| SIRPA | VEPH1 | 0.659386 | 3 | 0 | 3 |
| SIRPA | OLIG3 | 0.658713 | 3 | 0 | 3 |
| SIRPA | PEAR1 | 0.653925 | 3 | 0 | 3 |
| SIRPA | CDC25B | 0.650152 | 9 | 0 | 9 |
| SIRPA | SLC35B4 | 0.64978 | 4 | 0 | 3 |
| SIRPA | SNAI2 | 0.649203 | 9 | 0 | 8 |
| SIRPA | BCAT1 | 0.648886 | 4 | 0 | 4 |
For details and further investigation, click here