| Full name: spermine synthase | Alias Symbol: SPMSY|SpS|MRSR | ||
| Type: protein-coding gene | Cytoband: Xp22.11 | ||
| Entrez ID: 6611 | HGNC ID: HGNC:11123 | Ensembl Gene: ENSG00000102172 | OMIM ID: 300105 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SMS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMS | 6611 | 202043_s_at | 0.7992 | 0.0632 | |
| GSE20347 | SMS | 6611 | 202043_s_at | 0.8859 | 0.0025 | |
| GSE23400 | SMS | 6611 | 202043_s_at | 0.5819 | 0.0000 | |
| GSE26886 | SMS | 6611 | 202043_s_at | -0.4021 | 0.1543 | |
| GSE29001 | SMS | 6611 | 202043_s_at | 0.2298 | 0.5536 | |
| GSE38129 | SMS | 6611 | 202043_s_at | 0.7120 | 0.0008 | |
| GSE45670 | SMS | 6611 | 202043_s_at | 0.4288 | 0.0380 | |
| GSE53622 | SMS | 6611 | 29193 | 0.6268 | 0.0000 | |
| GSE53624 | SMS | 6611 | 29193 | 0.7773 | 0.0000 | |
| GSE63941 | SMS | 6611 | 202043_s_at | -0.2023 | 0.8078 | |
| GSE77861 | SMS | 6611 | 202043_s_at | 0.9502 | 0.0257 | |
| GSE97050 | SMS | A_21_P0010565 | 0.0177 | 0.9440 | ||
| SRP007169 | SMS | 6611 | RNAseq | -0.3053 | 0.5324 | |
| SRP008496 | SMS | 6611 | RNAseq | -0.2144 | 0.5300 | |
| SRP064894 | SMS | 6611 | RNAseq | 0.3248 | 0.0772 | |
| SRP133303 | SMS | 6611 | RNAseq | 1.2003 | 0.0009 | |
| SRP159526 | SMS | 6611 | RNAseq | 0.9710 | 0.0025 | |
| SRP193095 | SMS | 6611 | RNAseq | 0.4452 | 0.0160 | |
| SRP219564 | SMS | 6611 | RNAseq | 0.5637 | 0.1562 | |
| TCGA | SMS | 6611 | RNAseq | 0.1744 | 0.0156 |
Upregulated datasets: 1; Downregulated datasets: 0.
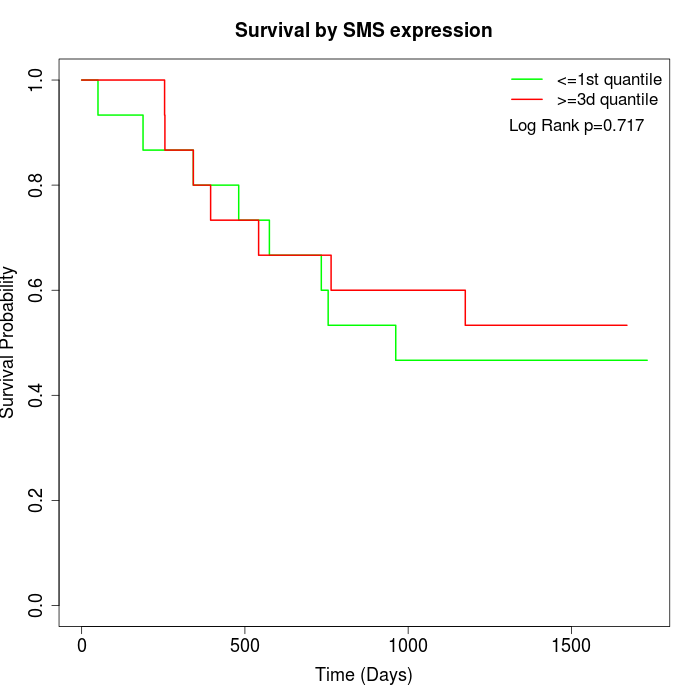
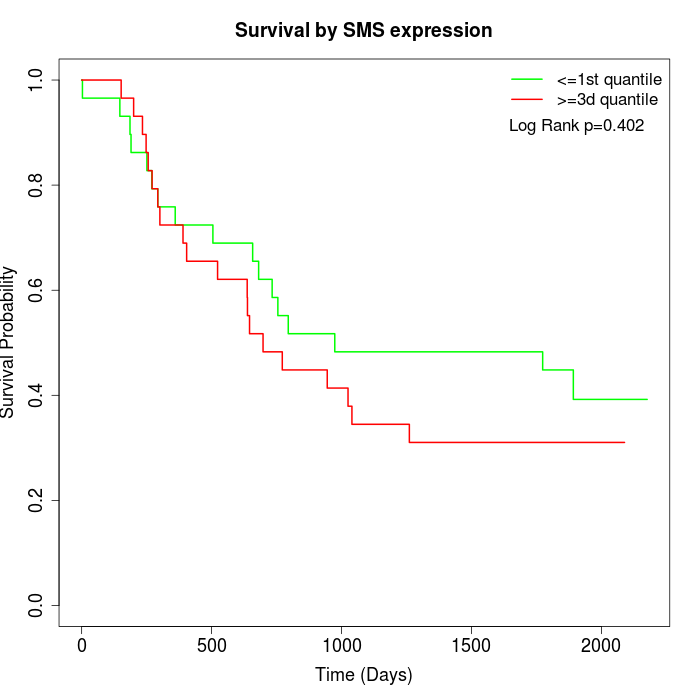
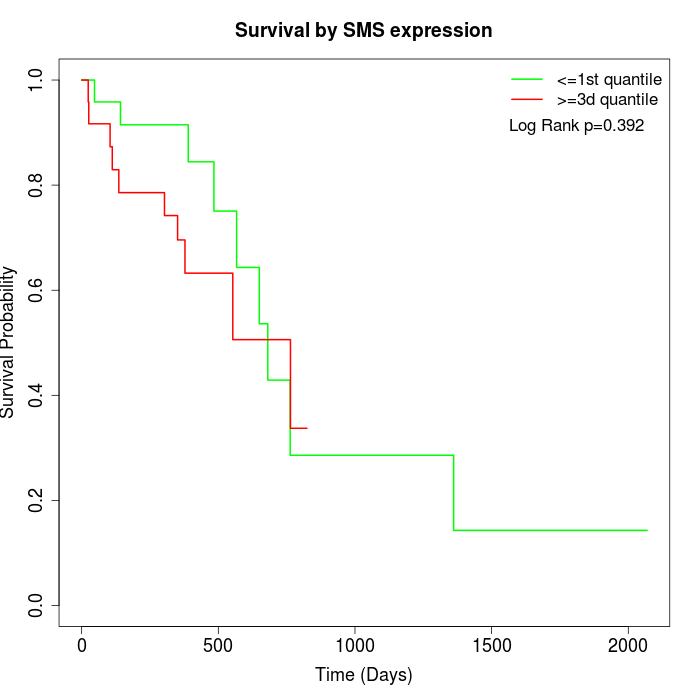
Survival by SMS expression:
Note: Click image to view full size file.
Copy number change of SMS:
No record found for this gene.
Somatic mutations of SMS:
Generating mutation plots.
Highly correlated genes for SMS:
Showing top 20/967 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMS | ZNF121 | 0.77578 | 3 | 0 | 3 |
| SMS | DUS3L | 0.771463 | 3 | 0 | 3 |
| SMS | NEK9 | 0.746345 | 3 | 0 | 3 |
| SMS | SPTBN1 | 0.732927 | 3 | 0 | 3 |
| SMS | RAVER1 | 0.7227 | 4 | 0 | 3 |
| SMS | FADS1 | 0.719774 | 3 | 0 | 3 |
| SMS | ZNF71 | 0.715303 | 3 | 0 | 3 |
| SMS | ZBTB2 | 0.711878 | 5 | 0 | 5 |
| SMS | MRPS5 | 0.707878 | 3 | 0 | 3 |
| SMS | MSL3 | 0.704328 | 4 | 0 | 3 |
| SMS | SUZ12 | 0.703749 | 4 | 0 | 4 |
| SMS | BCL7B | 0.703299 | 5 | 0 | 4 |
| SMS | CDCA2 | 0.703175 | 4 | 0 | 3 |
| SMS | TUBGCP2 | 0.698539 | 3 | 0 | 3 |
| SMS | TNKS | 0.696029 | 3 | 0 | 3 |
| SMS | TCF7L1 | 0.695733 | 3 | 0 | 3 |
| SMS | DAG1 | 0.69361 | 3 | 0 | 3 |
| SMS | GNG10 | 0.693063 | 3 | 0 | 3 |
| SMS | UTP23 | 0.690142 | 4 | 0 | 4 |
| SMS | TUBGCP4 | 0.690014 | 3 | 0 | 3 |
For details and further investigation, click here