| Full name: tankyrase | Alias Symbol: TIN1|TINF1|TNKS1|PARP-5a|PARP5A|pART5 | ||
| Type: protein-coding gene | Cytoband: 8p23.1 | ||
| Entrez ID: 8658 | HGNC ID: HGNC:11941 | Ensembl Gene: ENSG00000173273 | OMIM ID: 603303 |
| Drug and gene relationship at DGIdb | |||
Expression of TNKS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNKS | 8658 | 202561_at | 0.4063 | 0.4258 | |
| GSE20347 | TNKS | 8658 | 202561_at | 0.3393 | 0.1174 | |
| GSE23400 | TNKS | 8658 | 202561_at | 0.3000 | 0.0020 | |
| GSE26886 | TNKS | 8658 | 202561_at | 0.5403 | 0.0363 | |
| GSE29001 | TNKS | 8658 | 202561_at | 0.0889 | 0.7870 | |
| GSE38129 | TNKS | 8658 | 202561_at | 0.1178 | 0.5177 | |
| GSE45670 | TNKS | 8658 | 202561_at | -0.1833 | 0.3238 | |
| GSE53622 | TNKS | 8658 | 38044 | 0.0348 | 0.7580 | |
| GSE53624 | TNKS | 8658 | 38044 | -0.0018 | 0.9856 | |
| GSE63941 | TNKS | 8658 | 202561_at | -0.9969 | 0.1208 | |
| GSE77861 | TNKS | 8658 | 202561_at | 0.6450 | 0.1447 | |
| GSE97050 | TNKS | 8658 | A_32_P144596 | -0.0538 | 0.8828 | |
| SRP007169 | TNKS | 8658 | RNAseq | 0.6244 | 0.1445 | |
| SRP008496 | TNKS | 8658 | RNAseq | 0.6661 | 0.0029 | |
| SRP064894 | TNKS | 8658 | RNAseq | -0.0250 | 0.9113 | |
| SRP133303 | TNKS | 8658 | RNAseq | -0.0445 | 0.8102 | |
| SRP159526 | TNKS | 8658 | RNAseq | 0.1433 | 0.6340 | |
| SRP193095 | TNKS | 8658 | RNAseq | -0.0030 | 0.9744 | |
| SRP219564 | TNKS | 8658 | RNAseq | -0.0141 | 0.9504 | |
| TCGA | TNKS | 8658 | RNAseq | -0.0846 | 0.1244 |
Upregulated datasets: 0; Downregulated datasets: 0.
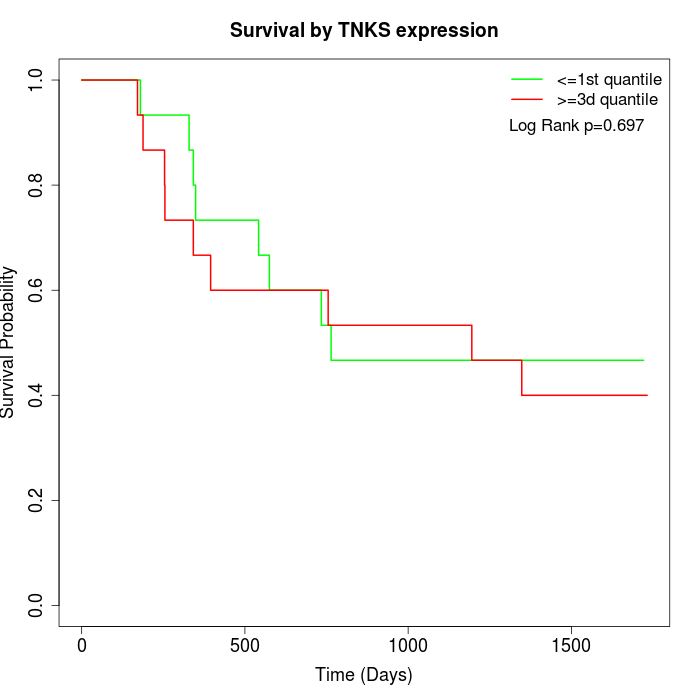
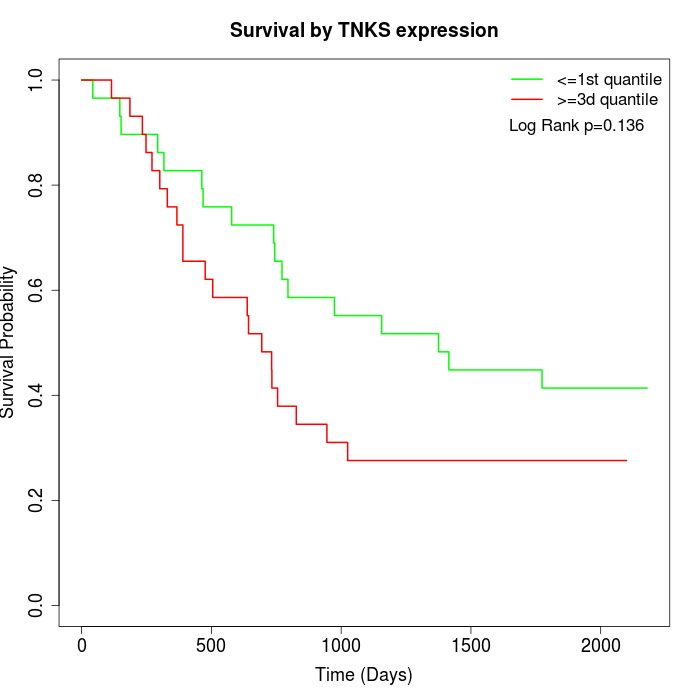
Survival by TNKS expression:
Note: Click image to view full size file.
Copy number change of TNKS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNKS | 8658 | 4 | 11 | 15 | |
| GSE20123 | TNKS | 8658 | 4 | 11 | 15 | |
| GSE43470 | TNKS | 8658 | 4 | 9 | 30 | |
| GSE46452 | TNKS | 8658 | 12 | 14 | 33 | |
| GSE47630 | TNKS | 8658 | 10 | 7 | 23 | |
| GSE54993 | TNKS | 8658 | 3 | 14 | 53 | |
| GSE54994 | TNKS | 8658 | 9 | 18 | 26 | |
| GSE60625 | TNKS | 8658 | 3 | 0 | 8 | |
| GSE74703 | TNKS | 8658 | 4 | 7 | 25 | |
| GSE74704 | TNKS | 8658 | 3 | 7 | 10 | |
| TCGA | TNKS | 8658 | 13 | 40 | 43 |
Total number of gains: 69; Total number of losses: 138; Total Number of normals: 281.
Somatic mutations of TNKS:
Generating mutation plots.
Highly correlated genes for TNKS:
Showing top 20/418 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNKS | ZDHHC7 | 0.752779 | 3 | 0 | 3 |
| TNKS | CLDN12 | 0.740409 | 4 | 0 | 4 |
| TNKS | GOLPH3 | 0.74019 | 3 | 0 | 3 |
| TNKS | SFT2D3 | 0.708294 | 3 | 0 | 3 |
| TNKS | SMS | 0.696029 | 3 | 0 | 3 |
| TNKS | USP37 | 0.686396 | 3 | 0 | 3 |
| TNKS | BRD9 | 0.685995 | 3 | 0 | 3 |
| TNKS | SRRT | 0.685323 | 3 | 0 | 3 |
| TNKS | LCLAT1 | 0.679432 | 4 | 0 | 3 |
| TNKS | LIG3 | 0.677988 | 4 | 0 | 4 |
| TNKS | SLC7A6 | 0.674232 | 4 | 0 | 3 |
| TNKS | LOXL1-AS1 | 0.66694 | 3 | 0 | 3 |
| TNKS | RICTOR | 0.665843 | 3 | 0 | 3 |
| TNKS | NRP2 | 0.664487 | 3 | 0 | 3 |
| TNKS | NPNT | 0.659703 | 3 | 0 | 3 |
| TNKS | KLHL5 | 0.65806 | 4 | 0 | 4 |
| TNKS | MAD2L2 | 0.646837 | 3 | 0 | 3 |
| TNKS | ASH2L | 0.642995 | 4 | 0 | 3 |
| TNKS | PDE7A | 0.642893 | 3 | 0 | 3 |
| TNKS | ZNF518B | 0.642393 | 3 | 0 | 3 |
For details and further investigation, click here