| Full name: signal peptidase complex subunit 1 | Alias Symbol: SPC12|HSPC033|YJR010C-A|SPC1 | ||
| Type: protein-coding gene | Cytoband: 3p21.1 | ||
| Entrez ID: 28972 | HGNC ID: HGNC:23401 | Ensembl Gene: ENSG00000114902 | OMIM ID: 610358 |
| Drug and gene relationship at DGIdb | |||
Expression of SPCS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPCS1 | 28972 | 217927_at | -0.2547 | 0.4059 | |
| GSE20347 | SPCS1 | 28972 | 217927_at | -0.6582 | 0.0001 | |
| GSE23400 | SPCS1 | 28972 | 217927_at | -0.2736 | 0.0000 | |
| GSE26886 | SPCS1 | 28972 | 217927_at | -1.1541 | 0.0000 | |
| GSE29001 | SPCS1 | 28972 | 217927_at | -0.3730 | 0.2250 | |
| GSE38129 | SPCS1 | 28972 | 217927_at | -0.3931 | 0.0083 | |
| GSE45670 | SPCS1 | 28972 | 217927_at | -0.2953 | 0.0046 | |
| GSE53622 | SPCS1 | 28972 | 46852 | -0.2909 | 0.0000 | |
| GSE53624 | SPCS1 | 28972 | 46852 | -0.4936 | 0.0000 | |
| GSE63941 | SPCS1 | 28972 | 217927_at | -0.3972 | 0.1496 | |
| GSE77861 | SPCS1 | 28972 | 217927_at | -0.8050 | 0.0016 | |
| GSE97050 | SPCS1 | 28972 | A_23_P29630 | 0.0148 | 0.9650 | |
| SRP007169 | SPCS1 | 28972 | RNAseq | -1.0999 | 0.0013 | |
| SRP008496 | SPCS1 | 28972 | RNAseq | -1.0572 | 0.0000 | |
| SRP064894 | SPCS1 | 28972 | RNAseq | -0.4559 | 0.0112 | |
| SRP133303 | SPCS1 | 28972 | RNAseq | -0.3562 | 0.0197 | |
| SRP159526 | SPCS1 | 28972 | RNAseq | -0.4269 | 0.0588 | |
| SRP193095 | SPCS1 | 28972 | RNAseq | -0.7618 | 0.0000 | |
| SRP219564 | SPCS1 | 28972 | RNAseq | -0.4933 | 0.0054 | |
| TCGA | SPCS1 | 28972 | RNAseq | -0.1787 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 3.
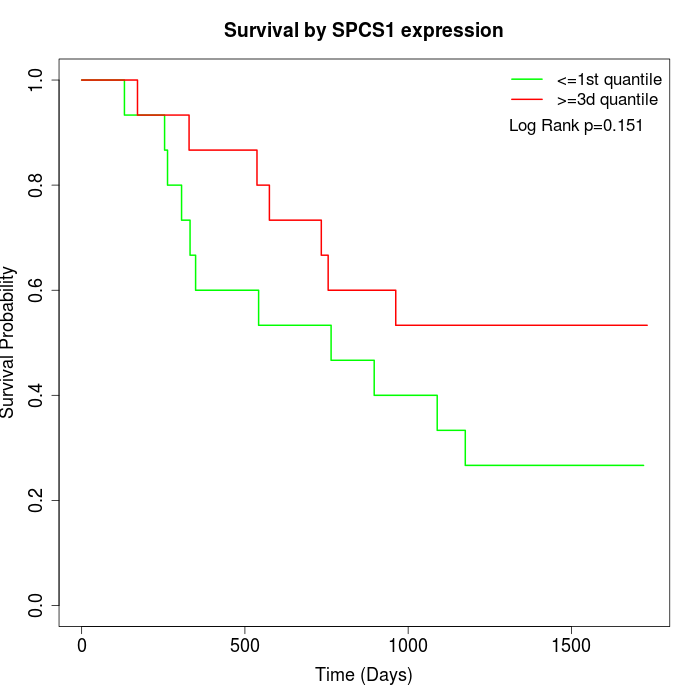
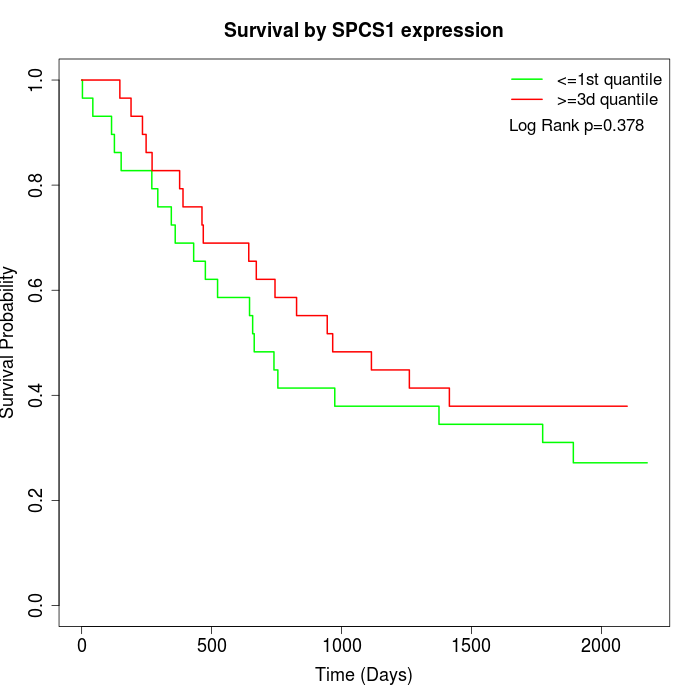
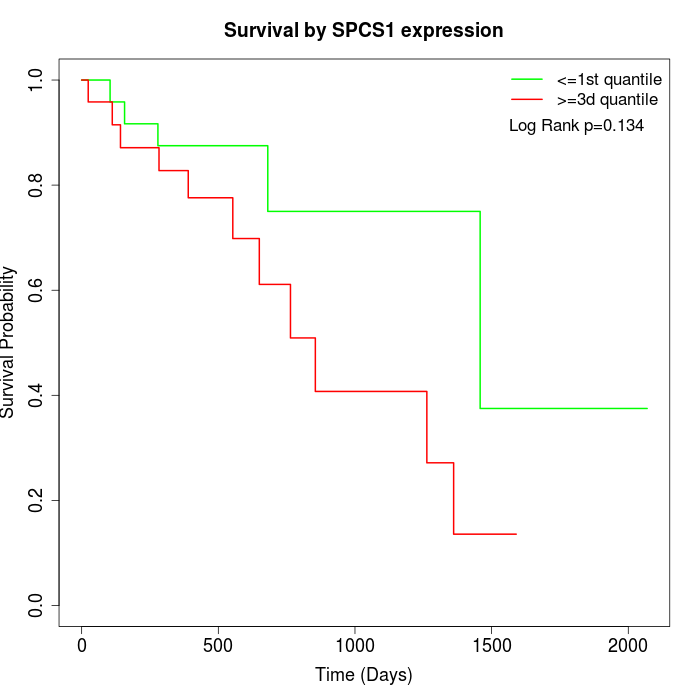
Survival by SPCS1 expression:
Note: Click image to view full size file.
Copy number change of SPCS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPCS1 | 28972 | 0 | 18 | 12 | |
| GSE20123 | SPCS1 | 28972 | 0 | 19 | 11 | |
| GSE43470 | SPCS1 | 28972 | 0 | 18 | 25 | |
| GSE46452 | SPCS1 | 28972 | 2 | 17 | 40 | |
| GSE47630 | SPCS1 | 28972 | 1 | 24 | 15 | |
| GSE54993 | SPCS1 | 28972 | 6 | 2 | 62 | |
| GSE54994 | SPCS1 | 28972 | 0 | 33 | 20 | |
| GSE60625 | SPCS1 | 28972 | 5 | 0 | 6 | |
| GSE74703 | SPCS1 | 28972 | 0 | 14 | 22 | |
| GSE74704 | SPCS1 | 28972 | 0 | 12 | 8 | |
| TCGA | SPCS1 | 28972 | 0 | 78 | 18 |
Total number of gains: 14; Total number of losses: 235; Total Number of normals: 239.
Somatic mutations of SPCS1:
Generating mutation plots.
Highly correlated genes for SPCS1:
Showing top 20/1394 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPCS1 | IMPACT | 0.786733 | 3 | 0 | 3 |
| SPCS1 | RHOA | 0.754925 | 13 | 0 | 12 |
| SPCS1 | DLG1 | 0.733427 | 3 | 0 | 3 |
| SPCS1 | TIFA | 0.725805 | 7 | 0 | 6 |
| SPCS1 | ST6GALNAC1 | 0.716671 | 6 | 0 | 6 |
| SPCS1 | RABAC1 | 0.715763 | 3 | 0 | 3 |
| SPCS1 | RASSF5 | 0.715008 | 6 | 0 | 6 |
| SPCS1 | ZNF436 | 0.712373 | 5 | 0 | 5 |
| SPCS1 | BRK1 | 0.711585 | 8 | 0 | 7 |
| SPCS1 | THAP2 | 0.710736 | 5 | 0 | 4 |
| SPCS1 | KLB | 0.709936 | 6 | 0 | 6 |
| SPCS1 | GMPPB | 0.705101 | 3 | 0 | 3 |
| SPCS1 | SNORA68 | 0.701711 | 4 | 0 | 4 |
| SPCS1 | GPR108 | 0.700555 | 3 | 0 | 3 |
| SPCS1 | MAN1A2 | 0.696683 | 3 | 0 | 3 |
| SPCS1 | FAM3D | 0.696125 | 6 | 0 | 5 |
| SPCS1 | PI4K2B | 0.695596 | 3 | 0 | 3 |
| SPCS1 | DCAF10 | 0.694445 | 6 | 0 | 5 |
| SPCS1 | DOCK8 | 0.693107 | 4 | 0 | 3 |
| SPCS1 | PLEKHA7 | 0.692213 | 7 | 0 | 7 |
For details and further investigation, click here