| Full name: spectrin alpha, non-erythrocytic 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q34.11 | ||
| Entrez ID: 6709 | HGNC ID: HGNC:11273 | Ensembl Gene: ENSG00000197694 | OMIM ID: 182810 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
SPTAN1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04210 | Apoptosis |
Expression of SPTAN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPTAN1 | 6709 | 208611_s_at | -0.0320 | 0.9269 | |
| GSE20347 | SPTAN1 | 6709 | 215235_at | -0.2058 | 0.0662 | |
| GSE23400 | SPTAN1 | 6709 | 208611_s_at | -0.2531 | 0.0000 | |
| GSE26886 | SPTAN1 | 6709 | 215235_at | 0.0518 | 0.8453 | |
| GSE29001 | SPTAN1 | 6709 | 215235_at | -0.0523 | 0.9617 | |
| GSE38129 | SPTAN1 | 6709 | 215235_at | -0.3751 | 0.0010 | |
| GSE45670 | SPTAN1 | 6709 | 215235_at | -0.2260 | 0.0481 | |
| GSE53622 | SPTAN1 | 6709 | 83936 | -0.5117 | 0.0000 | |
| GSE53624 | SPTAN1 | 6709 | 129986 | -0.1911 | 0.0000 | |
| GSE63941 | SPTAN1 | 6709 | 208611_s_at | 2.3041 | 0.0000 | |
| GSE77861 | SPTAN1 | 6709 | 208611_s_at | 0.1469 | 0.5764 | |
| GSE97050 | SPTAN1 | 6709 | A_33_P3373560 | -0.3555 | 0.1882 | |
| SRP007169 | SPTAN1 | 6709 | RNAseq | 0.2883 | 0.4048 | |
| SRP008496 | SPTAN1 | 6709 | RNAseq | 0.1544 | 0.5836 | |
| SRP064894 | SPTAN1 | 6709 | RNAseq | -0.1545 | 0.3296 | |
| SRP133303 | SPTAN1 | 6709 | RNAseq | -0.2537 | 0.0811 | |
| SRP159526 | SPTAN1 | 6709 | RNAseq | -0.2990 | 0.1399 | |
| SRP193095 | SPTAN1 | 6709 | RNAseq | -0.1852 | 0.0512 | |
| SRP219564 | SPTAN1 | 6709 | RNAseq | -0.5335 | 0.1095 | |
| TCGA | SPTAN1 | 6709 | RNAseq | -0.0963 | 0.0109 |
Upregulated datasets: 1; Downregulated datasets: 0.
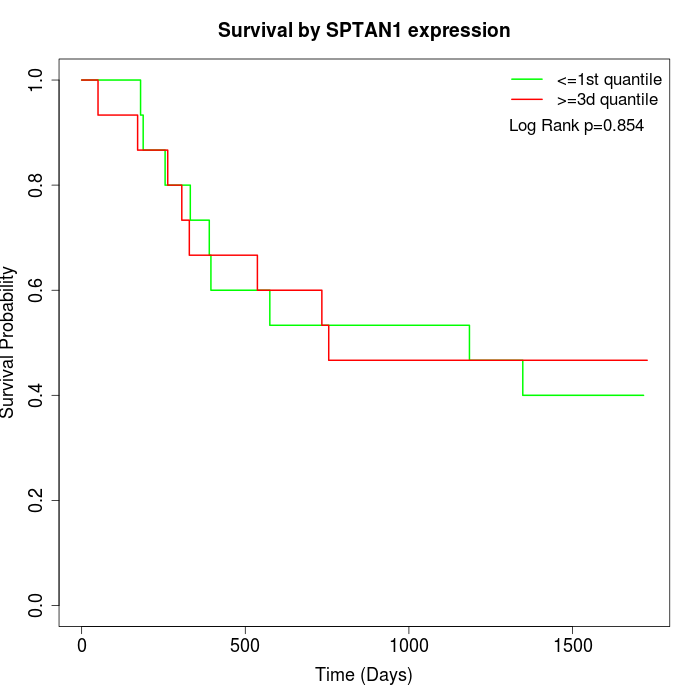
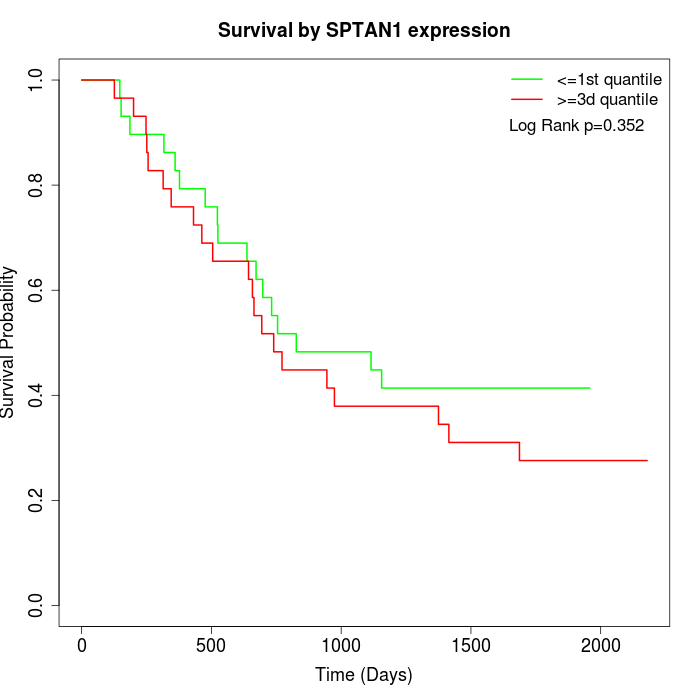
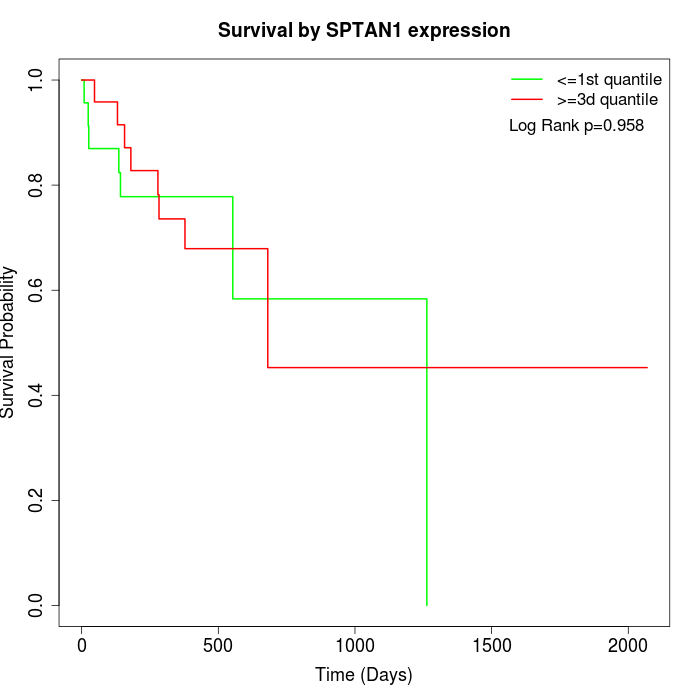
Survival by SPTAN1 expression:
Note: Click image to view full size file.
Copy number change of SPTAN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPTAN1 | 6709 | 6 | 9 | 15 | |
| GSE20123 | SPTAN1 | 6709 | 6 | 9 | 15 | |
| GSE43470 | SPTAN1 | 6709 | 5 | 8 | 30 | |
| GSE46452 | SPTAN1 | 6709 | 6 | 13 | 40 | |
| GSE47630 | SPTAN1 | 6709 | 3 | 16 | 21 | |
| GSE54993 | SPTAN1 | 6709 | 3 | 3 | 64 | |
| GSE54994 | SPTAN1 | 6709 | 11 | 8 | 34 | |
| GSE60625 | SPTAN1 | 6709 | 0 | 0 | 11 | |
| GSE74703 | SPTAN1 | 6709 | 5 | 6 | 25 | |
| GSE74704 | SPTAN1 | 6709 | 3 | 7 | 10 | |
| TCGA | SPTAN1 | 6709 | 27 | 24 | 45 |
Total number of gains: 75; Total number of losses: 103; Total Number of normals: 310.
Somatic mutations of SPTAN1:
Generating mutation plots.
Highly correlated genes for SPTAN1:
Showing top 20/392 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPTAN1 | MRPL38 | 0.822487 | 3 | 0 | 3 |
| SPTAN1 | ZNF362 | 0.811633 | 3 | 0 | 3 |
| SPTAN1 | ANP32B | 0.802334 | 3 | 0 | 3 |
| SPTAN1 | SF3A2 | 0.793833 | 3 | 0 | 3 |
| SPTAN1 | HHLA3 | 0.780262 | 4 | 0 | 4 |
| SPTAN1 | MDFI | 0.766851 | 3 | 0 | 3 |
| SPTAN1 | AAAS | 0.761496 | 5 | 0 | 5 |
| SPTAN1 | CCDC85C | 0.76 | 3 | 0 | 3 |
| SPTAN1 | ADAMTS10 | 0.758407 | 3 | 0 | 3 |
| SPTAN1 | COMT | 0.757026 | 3 | 0 | 3 |
| SPTAN1 | NCOR2 | 0.747033 | 3 | 0 | 3 |
| SPTAN1 | ZNF691 | 0.745868 | 3 | 0 | 3 |
| SPTAN1 | NAP1L4 | 0.745352 | 3 | 0 | 3 |
| SPTAN1 | ADRB3 | 0.744103 | 3 | 0 | 3 |
| SPTAN1 | MIF | 0.738636 | 3 | 0 | 3 |
| SPTAN1 | GGN | 0.737504 | 3 | 0 | 3 |
| SPTAN1 | DGKD | 0.737465 | 3 | 0 | 3 |
| SPTAN1 | ZNF408 | 0.733783 | 3 | 0 | 3 |
| SPTAN1 | PA2G4 | 0.726401 | 4 | 0 | 4 |
| SPTAN1 | AURKAIP1 | 0.722849 | 3 | 0 | 3 |
For details and further investigation, click here