| Full name: TGF-beta activated kinase 1 (MAP3K7) binding protein 2 | Alias Symbol: KIAA0733 | ||
| Type: protein-coding gene | Cytoband: 6q25.1 | ||
| Entrez ID: 23118 | HGNC ID: HGNC:17075 | Ensembl Gene: ENSG00000055208 | OMIM ID: 605101 |
| Drug and gene relationship at DGIdb | |||
TAB2 involved pathways:
Expression of TAB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TAB2 | 23118 | 212184_s_at | -0.3552 | 0.1074 | |
| GSE20347 | TAB2 | 23118 | 212184_s_at | -0.7077 | 0.0000 | |
| GSE23400 | TAB2 | 23118 | 210284_s_at | -0.4668 | 0.0000 | |
| GSE26886 | TAB2 | 23118 | 212184_s_at | -0.1946 | 0.4416 | |
| GSE29001 | TAB2 | 23118 | 212184_s_at | -0.5229 | 0.0992 | |
| GSE38129 | TAB2 | 23118 | 212184_s_at | -0.6903 | 0.0000 | |
| GSE45670 | TAB2 | 23118 | 212184_s_at | -0.5037 | 0.0004 | |
| GSE53622 | TAB2 | 23118 | 55349 | -0.2828 | 0.0000 | |
| GSE53624 | TAB2 | 23118 | 65520 | -0.5146 | 0.0000 | |
| GSE63941 | TAB2 | 23118 | 212184_s_at | -0.4494 | 0.2294 | |
| GSE77861 | TAB2 | 23118 | 212184_s_at | -0.6476 | 0.0418 | |
| GSE97050 | TAB2 | 23118 | A_33_P3404331 | -0.2240 | 0.2565 | |
| SRP007169 | TAB2 | 23118 | RNAseq | -0.9335 | 0.0419 | |
| SRP008496 | TAB2 | 23118 | RNAseq | -0.6678 | 0.0148 | |
| SRP064894 | TAB2 | 23118 | RNAseq | -0.7526 | 0.0001 | |
| SRP133303 | TAB2 | 23118 | RNAseq | -0.1303 | 0.3733 | |
| SRP159526 | TAB2 | 23118 | RNAseq | -0.6366 | 0.0073 | |
| SRP193095 | TAB2 | 23118 | RNAseq | -0.6838 | 0.0000 | |
| SRP219564 | TAB2 | 23118 | RNAseq | -0.6509 | 0.0757 | |
| TCGA | TAB2 | 23118 | RNAseq | -0.0816 | 0.0609 |
Upregulated datasets: 0; Downregulated datasets: 0.
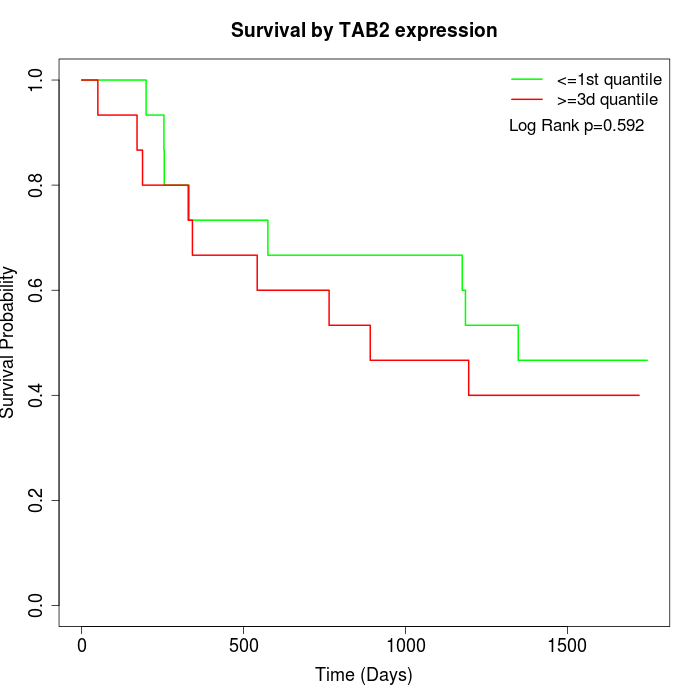
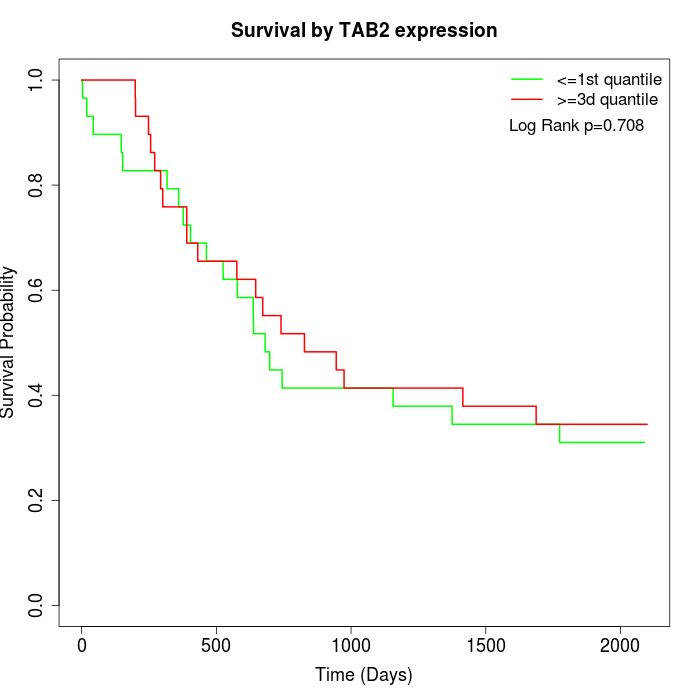
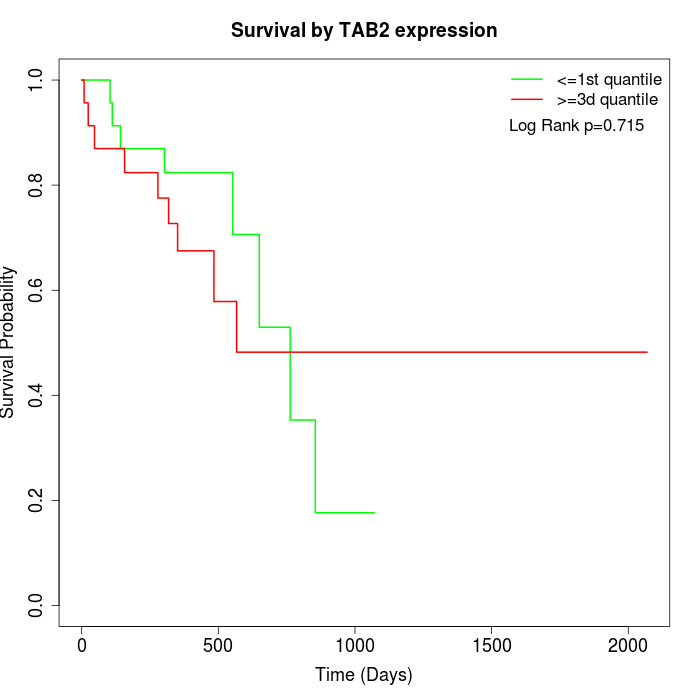
Survival by TAB2 expression:
Note: Click image to view full size file.
Copy number change of TAB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TAB2 | 23118 | 2 | 4 | 24 | |
| GSE20123 | TAB2 | 23118 | 1 | 3 | 26 | |
| GSE43470 | TAB2 | 23118 | 4 | 0 | 39 | |
| GSE46452 | TAB2 | 23118 | 3 | 10 | 46 | |
| GSE47630 | TAB2 | 23118 | 9 | 4 | 27 | |
| GSE54993 | TAB2 | 23118 | 3 | 2 | 65 | |
| GSE54994 | TAB2 | 23118 | 8 | 8 | 37 | |
| GSE60625 | TAB2 | 23118 | 0 | 1 | 10 | |
| GSE74703 | TAB2 | 23118 | 4 | 0 | 32 | |
| GSE74704 | TAB2 | 23118 | 0 | 1 | 19 | |
| TCGA | TAB2 | 23118 | 12 | 21 | 63 |
Total number of gains: 46; Total number of losses: 54; Total Number of normals: 388.
Somatic mutations of TAB2:
Generating mutation plots.
Highly correlated genes for TAB2:
Showing top 20/1211 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TAB2 | SERAC1 | 0.724587 | 4 | 0 | 4 |
| TAB2 | SNORA68 | 0.707633 | 3 | 0 | 3 |
| TAB2 | ZRANB1 | 0.704197 | 5 | 0 | 4 |
| TAB2 | C5orf66-AS1 | 0.697335 | 3 | 0 | 3 |
| TAB2 | SCRN3 | 0.688296 | 3 | 0 | 3 |
| TAB2 | SLK | 0.686058 | 9 | 0 | 9 |
| TAB2 | SNX3 | 0.68475 | 12 | 0 | 11 |
| TAB2 | RBMS3 | 0.680467 | 4 | 0 | 3 |
| TAB2 | UGP2 | 0.67553 | 9 | 0 | 8 |
| TAB2 | GAB1 | 0.675045 | 8 | 0 | 8 |
| TAB2 | TMEM59 | 0.674725 | 10 | 0 | 9 |
| TAB2 | STX7 | 0.674413 | 10 | 0 | 9 |
| TAB2 | SFTA2 | 0.672446 | 3 | 0 | 3 |
| TAB2 | PFDN5 | 0.671393 | 8 | 0 | 7 |
| TAB2 | RIOK3 | 0.669257 | 6 | 0 | 6 |
| TAB2 | CITED2 | 0.665854 | 11 | 0 | 8 |
| TAB2 | RAP1A | 0.662567 | 12 | 0 | 8 |
| TAB2 | NEK4 | 0.661879 | 3 | 0 | 3 |
| TAB2 | PINK1 | 0.661497 | 8 | 0 | 7 |
| TAB2 | SERINC1 | 0.660597 | 12 | 0 | 8 |
For details and further investigation, click here