| Full name: transcription factor 12 | Alias Symbol: HEB|HTF4|HsT17266|bHLHb20|p64 | ||
| Type: protein-coding gene | Cytoband: 15q21.3 | ||
| Entrez ID: 6938 | HGNC ID: HGNC:11623 | Ensembl Gene: ENSG00000140262 | OMIM ID: 600480 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TCF12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCF12 | 6938 | 208986_at | -0.1387 | 0.7526 | |
| GSE20347 | TCF12 | 6938 | 208986_at | 0.1085 | 0.5838 | |
| GSE23400 | TCF12 | 6938 | 208986_at | 0.1698 | 0.0339 | |
| GSE26886 | TCF12 | 6938 | 208986_at | 1.1074 | 0.0000 | |
| GSE29001 | TCF12 | 6938 | 208986_at | 0.1366 | 0.7239 | |
| GSE38129 | TCF12 | 6938 | 208986_at | 0.0969 | 0.4518 | |
| GSE45670 | TCF12 | 6938 | 208986_at | -0.2662 | 0.1439 | |
| GSE53622 | TCF12 | 6938 | 94023 | 0.1719 | 0.0219 | |
| GSE53624 | TCF12 | 6938 | 94023 | 0.0338 | 0.7971 | |
| GSE63941 | TCF12 | 6938 | 208986_at | -0.4138 | 0.3022 | |
| GSE77861 | TCF12 | 6938 | 208986_at | 0.1391 | 0.6325 | |
| GSE97050 | TCF12 | 6938 | A_24_P82142 | 0.4253 | 0.2379 | |
| SRP007169 | TCF12 | 6938 | RNAseq | 0.0646 | 0.8291 | |
| SRP008496 | TCF12 | 6938 | RNAseq | 0.2574 | 0.2362 | |
| SRP064894 | TCF12 | 6938 | RNAseq | 0.0389 | 0.8745 | |
| SRP133303 | TCF12 | 6938 | RNAseq | 0.3519 | 0.0123 | |
| SRP159526 | TCF12 | 6938 | RNAseq | 0.0843 | 0.6890 | |
| SRP193095 | TCF12 | 6938 | RNAseq | 0.2534 | 0.0354 | |
| SRP219564 | TCF12 | 6938 | RNAseq | 0.0535 | 0.8671 | |
| TCGA | TCF12 | 6938 | RNAseq | -0.0502 | 0.2534 |
Upregulated datasets: 1; Downregulated datasets: 0.
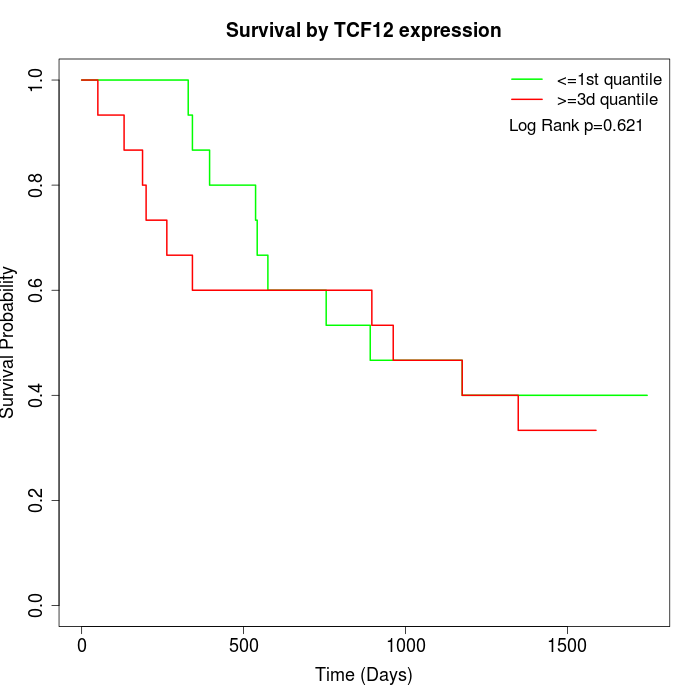
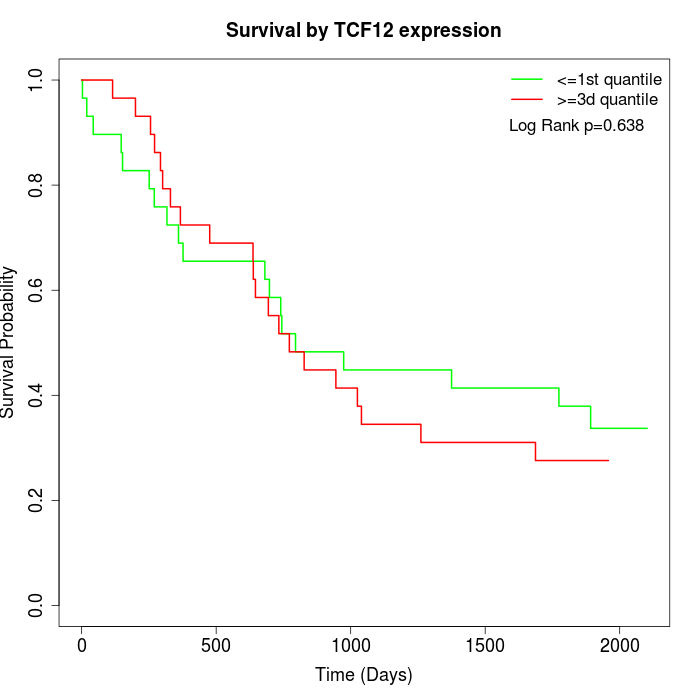
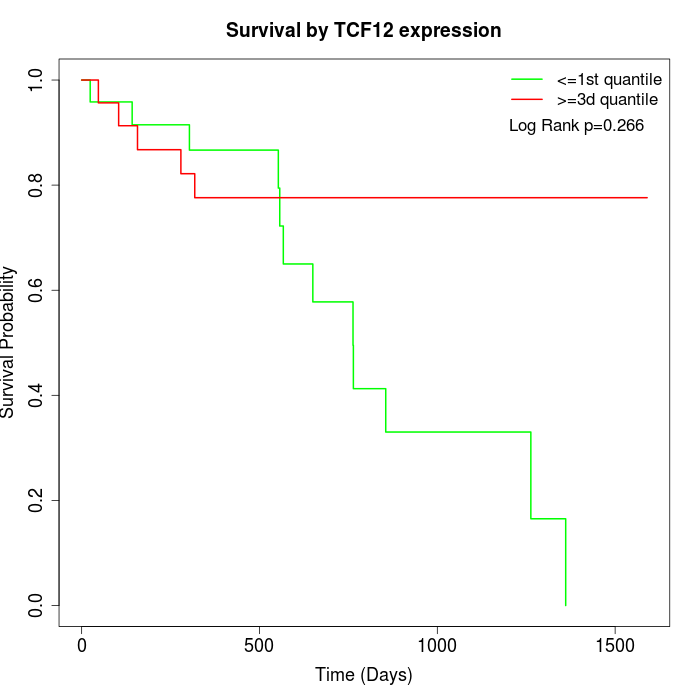
Survival by TCF12 expression:
Note: Click image to view full size file.
Copy number change of TCF12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TCF12 | 6938 | 6 | 3 | 21 | |
| GSE20123 | TCF12 | 6938 | 6 | 3 | 21 | |
| GSE43470 | TCF12 | 6938 | 4 | 4 | 35 | |
| GSE46452 | TCF12 | 6938 | 3 | 7 | 49 | |
| GSE47630 | TCF12 | 6938 | 8 | 10 | 22 | |
| GSE54993 | TCF12 | 6938 | 5 | 6 | 59 | |
| GSE54994 | TCF12 | 6938 | 6 | 8 | 39 | |
| GSE60625 | TCF12 | 6938 | 4 | 0 | 7 | |
| GSE74703 | TCF12 | 6938 | 4 | 3 | 29 | |
| GSE74704 | TCF12 | 6938 | 2 | 3 | 15 | |
| TCGA | TCF12 | 6938 | 10 | 17 | 69 |
Total number of gains: 58; Total number of losses: 64; Total Number of normals: 366.
Somatic mutations of TCF12:
Generating mutation plots.
Highly correlated genes for TCF12:
Showing top 20/628 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCF12 | PSMD3 | 0.770136 | 3 | 0 | 3 |
| TCF12 | TOPBP1 | 0.764012 | 3 | 0 | 3 |
| TCF12 | HIBADH | 0.757415 | 3 | 0 | 3 |
| TCF12 | SBF2 | 0.74969 | 3 | 0 | 3 |
| TCF12 | ERVW-1 | 0.745292 | 3 | 0 | 3 |
| TCF12 | LRRC23 | 0.734062 | 3 | 0 | 3 |
| TCF12 | GLO1 | 0.724782 | 4 | 0 | 4 |
| TCF12 | CCDC66 | 0.718377 | 3 | 0 | 3 |
| TCF12 | ARHGAP42 | 0.713338 | 3 | 0 | 3 |
| TCF12 | CWF19L2 | 0.710729 | 4 | 0 | 4 |
| TCF12 | MXRA7 | 0.707227 | 5 | 0 | 4 |
| TCF12 | COPZ2 | 0.706722 | 3 | 0 | 3 |
| TCF12 | WTAP | 0.706131 | 3 | 0 | 3 |
| TCF12 | LMBRD2 | 0.704799 | 3 | 0 | 3 |
| TCF12 | UBR3 | 0.702578 | 3 | 0 | 3 |
| TCF12 | RBM4 | 0.700213 | 3 | 0 | 3 |
| TCF12 | MRPL24 | 0.698699 | 3 | 0 | 3 |
| TCF12 | ZNF689 | 0.693279 | 3 | 0 | 3 |
| TCF12 | FAF2 | 0.692321 | 4 | 0 | 4 |
| TCF12 | SHPRH | 0.692308 | 3 | 0 | 3 |
For details and further investigation, click here