| Full name: WT1 associated protein | Alias Symbol: KIAA0105|MGC3925|Mum2 | ||
| Type: protein-coding gene | Cytoband: 6q25.3 | ||
| Entrez ID: 9589 | HGNC ID: HGNC:16846 | Ensembl Gene: ENSG00000146457 | OMIM ID: 605442 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of WTAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | WTAP | 9589 | 203137_at | 0.2719 | 0.6027 | |
| GSE20347 | WTAP | 9589 | 203137_at | -0.1317 | 0.5366 | |
| GSE23400 | WTAP | 9589 | 203137_at | 0.0730 | 0.4595 | |
| GSE26886 | WTAP | 9589 | 229630_s_at | -0.4090 | 0.0473 | |
| GSE29001 | WTAP | 9589 | 203137_at | -0.1629 | 0.7380 | |
| GSE38129 | WTAP | 9589 | 203137_at | 0.0668 | 0.6705 | |
| GSE45670 | WTAP | 9589 | 229630_s_at | 0.1245 | 0.2197 | |
| GSE53622 | WTAP | 9589 | 65997 | -0.1879 | 0.0159 | |
| GSE53624 | WTAP | 9589 | 65997 | -0.1980 | 0.0019 | |
| GSE63941 | WTAP | 9589 | 229630_s_at | 0.9604 | 0.0049 | |
| GSE77861 | WTAP | 9589 | 227621_at | 0.5055 | 0.0734 | |
| GSE97050 | WTAP | 9589 | A_33_P3329098 | 0.4215 | 0.1623 | |
| SRP007169 | WTAP | 9589 | RNAseq | -0.4982 | 0.0927 | |
| SRP008496 | WTAP | 9589 | RNAseq | -0.2939 | 0.1871 | |
| SRP064894 | WTAP | 9589 | RNAseq | 0.0990 | 0.5658 | |
| SRP133303 | WTAP | 9589 | RNAseq | 0.3177 | 0.0475 | |
| SRP159526 | WTAP | 9589 | RNAseq | -0.1302 | 0.5672 | |
| SRP193095 | WTAP | 9589 | RNAseq | -0.0143 | 0.8802 | |
| SRP219564 | WTAP | 9589 | RNAseq | 0.1715 | 0.5678 | |
| TCGA | WTAP | 9589 | RNAseq | 0.0574 | 0.2128 |
Upregulated datasets: 0; Downregulated datasets: 0.
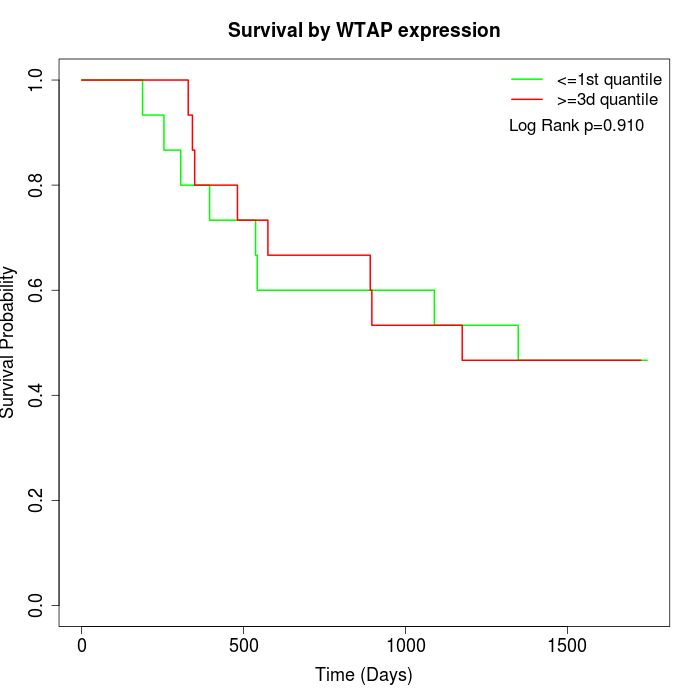
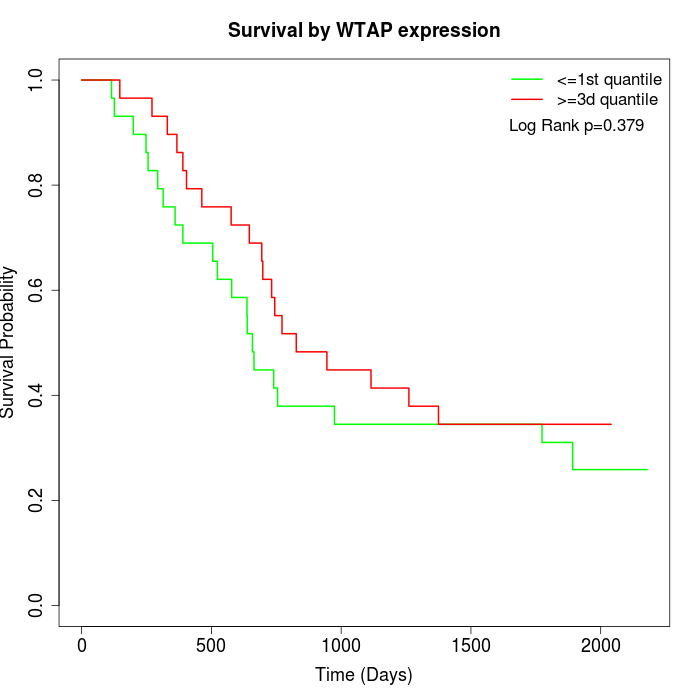
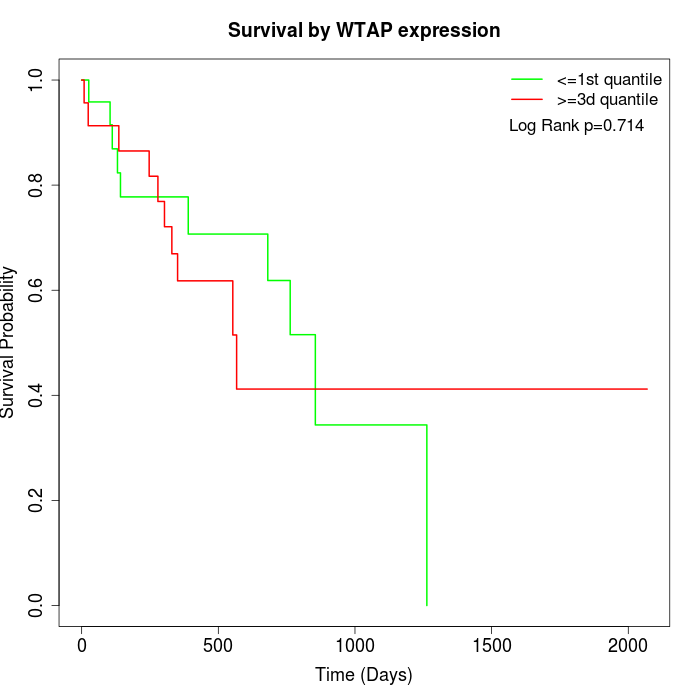
Survival by WTAP expression:
Note: Click image to view full size file.
Copy number change of WTAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | WTAP | 9589 | 1 | 5 | 24 | |
| GSE20123 | WTAP | 9589 | 1 | 4 | 25 | |
| GSE43470 | WTAP | 9589 | 4 | 0 | 39 | |
| GSE46452 | WTAP | 9589 | 3 | 10 | 46 | |
| GSE47630 | WTAP | 9589 | 9 | 4 | 27 | |
| GSE54993 | WTAP | 9589 | 3 | 2 | 65 | |
| GSE54994 | WTAP | 9589 | 8 | 8 | 37 | |
| GSE60625 | WTAP | 9589 | 0 | 1 | 10 | |
| GSE74703 | WTAP | 9589 | 4 | 0 | 32 | |
| GSE74704 | WTAP | 9589 | 0 | 2 | 18 | |
| TCGA | WTAP | 9589 | 13 | 22 | 61 |
Total number of gains: 46; Total number of losses: 58; Total Number of normals: 384.
Somatic mutations of WTAP:
Generating mutation plots.
Highly correlated genes for WTAP:
Showing top 20/435 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| WTAP | EXOC6 | 0.814046 | 3 | 0 | 3 |
| WTAP | EEFSEC | 0.807176 | 3 | 0 | 3 |
| WTAP | TSFM | 0.800396 | 3 | 0 | 3 |
| WTAP | ZNF292 | 0.769931 | 3 | 0 | 3 |
| WTAP | KANSL2 | 0.767855 | 5 | 0 | 5 |
| WTAP | TBPL1 | 0.749808 | 6 | 0 | 6 |
| WTAP | MMRN2 | 0.746734 | 3 | 0 | 3 |
| WTAP | GPR160 | 0.746514 | 3 | 0 | 3 |
| WTAP | TADA1 | 0.736743 | 3 | 0 | 3 |
| WTAP | EMG1 | 0.732033 | 3 | 0 | 3 |
| WTAP | CCNT1 | 0.724557 | 3 | 0 | 3 |
| WTAP | STK4 | 0.722192 | 3 | 0 | 3 |
| WTAP | NNT | 0.720714 | 3 | 0 | 3 |
| WTAP | PSME4 | 0.718374 | 3 | 0 | 3 |
| WTAP | RSL24D1 | 0.716923 | 5 | 0 | 5 |
| WTAP | CREB1 | 0.715138 | 4 | 0 | 4 |
| WTAP | RNF166 | 0.710837 | 3 | 0 | 3 |
| WTAP | TCF12 | 0.706131 | 3 | 0 | 3 |
| WTAP | RPP38 | 0.70235 | 4 | 0 | 4 |
| WTAP | ELK4 | 0.702161 | 3 | 0 | 3 |
For details and further investigation, click here