| Full name: TNF superfamily member 10 | Alias Symbol: TRAIL|Apo-2L|TL2|CD253 | ||
| Type: protein-coding gene | Cytoband: 3q26 | ||
| Entrez ID: 8743 | HGNC ID: HGNC:11925 | Ensembl Gene: ENSG00000121858 | OMIM ID: 603598 |
| Drug and gene relationship at DGIdb | |||
TNFSF10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04210 | Apoptosis | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa05162 | Measles |
Expression of TNFSF10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNFSF10 | 8743 | 202688_at | 0.1908 | 0.8264 | |
| GSE20347 | TNFSF10 | 8743 | 202688_at | 0.6817 | 0.0102 | |
| GSE23400 | TNFSF10 | 8743 | 202688_at | 0.7643 | 0.0000 | |
| GSE26886 | TNFSF10 | 8743 | 202688_at | 0.8520 | 0.0210 | |
| GSE29001 | TNFSF10 | 8743 | 214329_x_at | 0.7061 | 0.0334 | |
| GSE38129 | TNFSF10 | 8743 | 202688_at | 0.8019 | 0.0049 | |
| GSE45670 | TNFSF10 | 8743 | 202687_s_at | 1.0662 | 0.0011 | |
| GSE53622 | TNFSF10 | 8743 | 5418 | 1.0978 | 0.0000 | |
| GSE53624 | TNFSF10 | 8743 | 5418 | 0.9725 | 0.0000 | |
| GSE63941 | TNFSF10 | 8743 | 202688_at | 3.4334 | 0.0802 | |
| GSE77861 | TNFSF10 | 8743 | 202688_at | -0.2524 | 0.7646 | |
| GSE97050 | TNFSF10 | 8743 | A_33_P3226810 | 1.8177 | 0.0555 | |
| SRP007169 | TNFSF10 | 8743 | RNAseq | -0.3601 | 0.5736 | |
| SRP008496 | TNFSF10 | 8743 | RNAseq | -0.1524 | 0.7460 | |
| SRP064894 | TNFSF10 | 8743 | RNAseq | 1.0139 | 0.0005 | |
| SRP133303 | TNFSF10 | 8743 | RNAseq | 1.4935 | 0.0004 | |
| SRP159526 | TNFSF10 | 8743 | RNAseq | -0.0680 | 0.9314 | |
| SRP193095 | TNFSF10 | 8743 | RNAseq | 0.4634 | 0.0682 | |
| SRP219564 | TNFSF10 | 8743 | RNAseq | 0.8535 | 0.3122 | |
| TCGA | TNFSF10 | 8743 | RNAseq | 0.7213 | 0.0000 |
Upregulated datasets: 4; Downregulated datasets: 0.
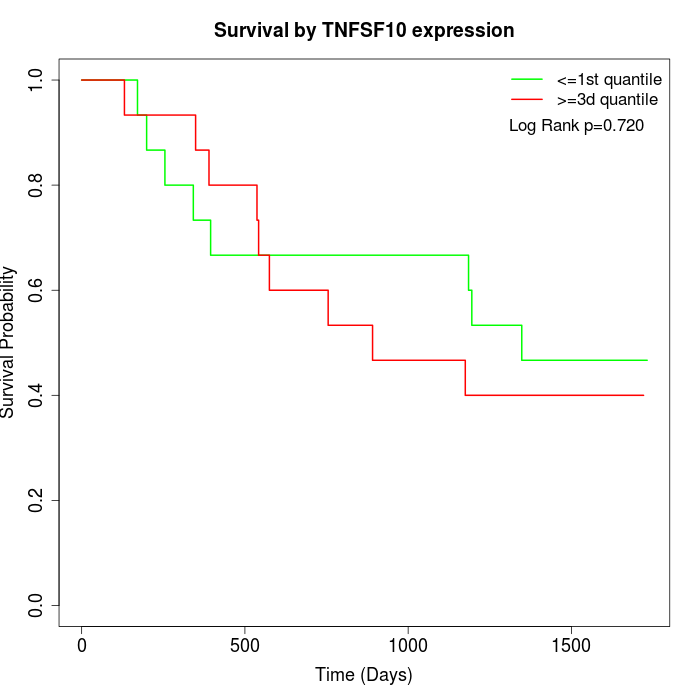
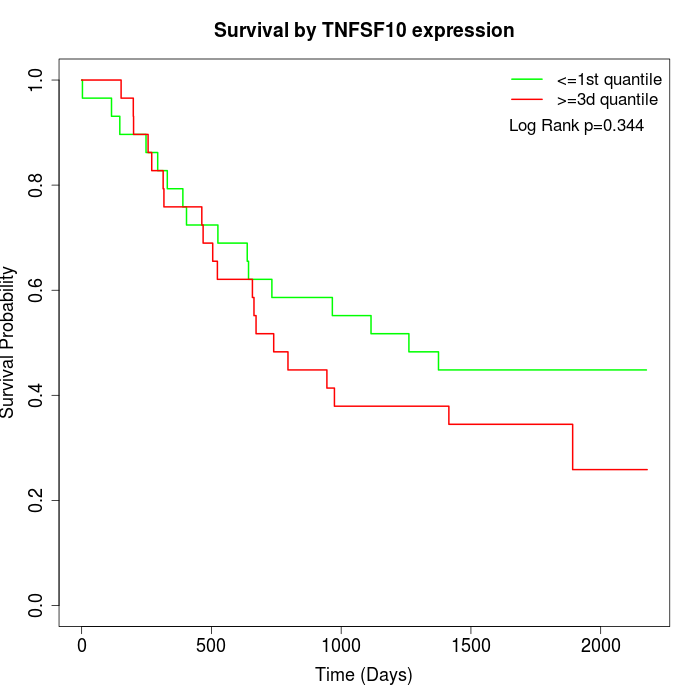
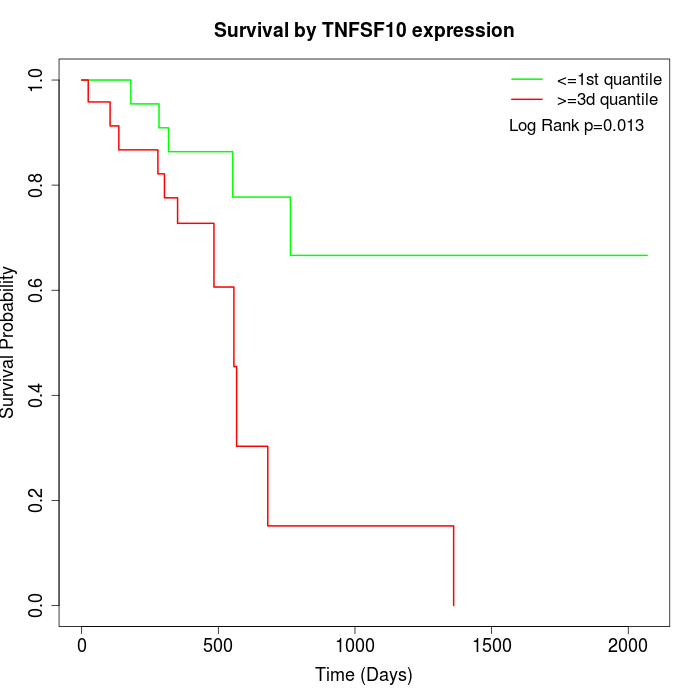
Survival by TNFSF10 expression:
Note: Click image to view full size file.
Copy number change of TNFSF10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNFSF10 | 8743 | 24 | 0 | 6 | |
| GSE20123 | TNFSF10 | 8743 | 24 | 0 | 6 | |
| GSE43470 | TNFSF10 | 8743 | 23 | 0 | 20 | |
| GSE46452 | TNFSF10 | 8743 | 19 | 1 | 39 | |
| GSE47630 | TNFSF10 | 8743 | 27 | 2 | 11 | |
| GSE54993 | TNFSF10 | 8743 | 1 | 15 | 54 | |
| GSE54994 | TNFSF10 | 8743 | 41 | 0 | 12 | |
| GSE60625 | TNFSF10 | 8743 | 0 | 6 | 5 | |
| GSE74703 | TNFSF10 | 8743 | 19 | 0 | 17 | |
| GSE74704 | TNFSF10 | 8743 | 16 | 0 | 4 | |
| TCGA | TNFSF10 | 8743 | 80 | 0 | 16 |
Total number of gains: 274; Total number of losses: 24; Total Number of normals: 190.
Somatic mutations of TNFSF10:
Generating mutation plots.
Highly correlated genes for TNFSF10:
Showing top 20/242 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNFSF10 | ITGAL | 0.753821 | 3 | 0 | 3 |
| TNFSF10 | VMP1 | 0.689069 | 3 | 0 | 3 |
| TNFSF10 | UBA2 | 0.687466 | 3 | 0 | 3 |
| TNFSF10 | DCBLD1 | 0.670299 | 3 | 0 | 3 |
| TNFSF10 | SLC12A7 | 0.63946 | 3 | 0 | 3 |
| TNFSF10 | DTX3L | 0.632991 | 9 | 0 | 8 |
| TNFSF10 | TMC8 | 0.622559 | 3 | 0 | 3 |
| TNFSF10 | CALML5 | 0.611979 | 4 | 0 | 3 |
| TNFSF10 | LIMK2 | 0.60737 | 4 | 0 | 4 |
| TNFSF10 | MDK | 0.605682 | 7 | 0 | 6 |
| TNFSF10 | IL15 | 0.6045 | 6 | 0 | 5 |
| TNFSF10 | RAP1GAP2 | 0.604192 | 3 | 0 | 3 |
| TNFSF10 | STAT2 | 0.603384 | 4 | 0 | 3 |
| TNFSF10 | IL10RA | 0.602929 | 3 | 0 | 3 |
| TNFSF10 | LAMB3 | 0.602026 | 9 | 0 | 7 |
| TNFSF10 | WDR66 | 0.601411 | 6 | 0 | 5 |
| TNFSF10 | USP41 | 0.60098 | 3 | 0 | 3 |
| TNFSF10 | CEP63 | 0.598087 | 3 | 0 | 3 |
| TNFSF10 | MAP3K5 | 0.591907 | 3 | 0 | 3 |
| TNFSF10 | GRTP1 | 0.589162 | 3 | 0 | 3 |
For details and further investigation, click here