| Full name: TRAM2 antisense RNA 1 (head to head) | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 6p12.2 | ||
| Entrez ID: 401264 | HGNC ID: HGNC:48663 | Ensembl Gene: ENSG00000225791 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TRAM2-AS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRAM2-AS1 | 401264 | 227298_at | -0.2473 | 0.5780 | |
| GSE26886 | TRAM2-AS1 | 401264 | 227298_at | 0.7607 | 0.0039 | |
| GSE45670 | TRAM2-AS1 | 401264 | 227298_at | -0.3025 | 0.1336 | |
| GSE53622 | TRAM2-AS1 | 401264 | 93533 | -0.0892 | 0.4413 | |
| GSE53624 | TRAM2-AS1 | 401264 | 93533 | 0.2587 | 0.0085 | |
| GSE63941 | TRAM2-AS1 | 401264 | 227298_at | -1.3114 | 0.0297 | |
| GSE77861 | TRAM2-AS1 | 401264 | 227298_at | 0.1920 | 0.5263 | |
| SRP064894 | TRAM2-AS1 | 401264 | RNAseq | 0.3469 | 0.1653 | |
| SRP133303 | TRAM2-AS1 | 401264 | RNAseq | 0.2381 | 0.1586 | |
| SRP159526 | TRAM2-AS1 | 401264 | RNAseq | 0.7570 | 0.0080 | |
| SRP193095 | TRAM2-AS1 | 401264 | RNAseq | 0.3314 | 0.1406 | |
| SRP219564 | TRAM2-AS1 | 401264 | RNAseq | 0.4473 | 0.4196 |
Upregulated datasets: 0; Downregulated datasets: 1.
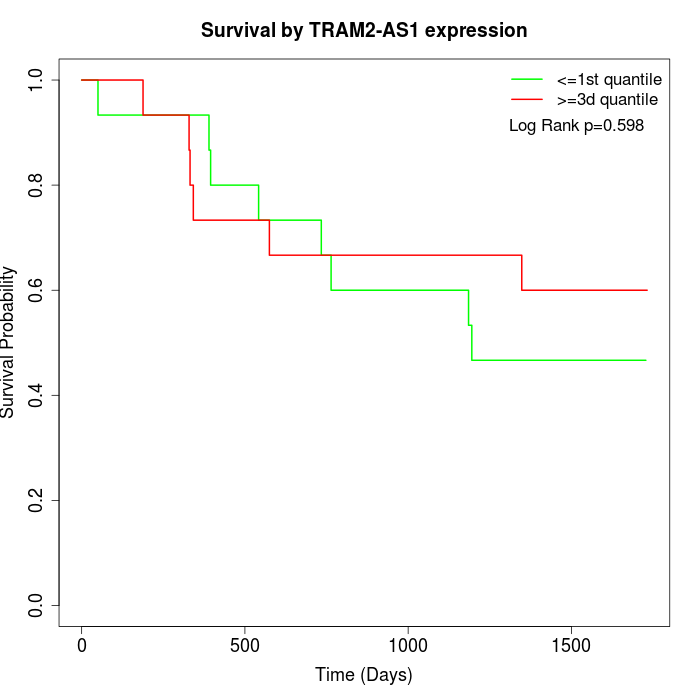
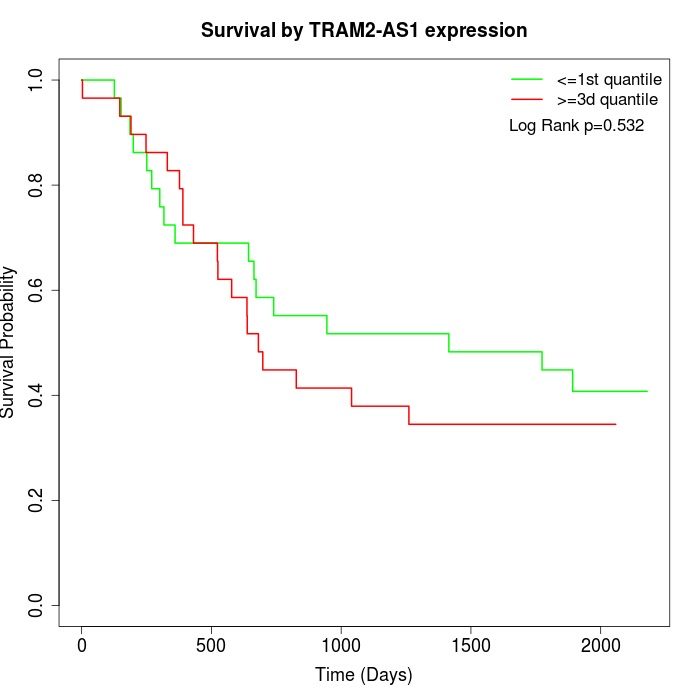
Survival by TRAM2-AS1 expression:
Note: Click image to view full size file.
Copy number change of TRAM2-AS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRAM2-AS1 | 401264 | 7 | 2 | 21 | |
| GSE20123 | TRAM2-AS1 | 401264 | 7 | 2 | 21 | |
| GSE43470 | TRAM2-AS1 | 401264 | 5 | 0 | 38 | |
| GSE46452 | TRAM2-AS1 | 401264 | 2 | 10 | 47 | |
| GSE47630 | TRAM2-AS1 | 401264 | 7 | 5 | 28 | |
| GSE54993 | TRAM2-AS1 | 401264 | 3 | 1 | 66 | |
| GSE54994 | TRAM2-AS1 | 401264 | 8 | 4 | 41 | |
| GSE60625 | TRAM2-AS1 | 401264 | 0 | 3 | 8 | |
| GSE74703 | TRAM2-AS1 | 401264 | 5 | 0 | 31 | |
| GSE74704 | TRAM2-AS1 | 401264 | 3 | 1 | 16 |
Total number of gains: 47; Total number of losses: 28; Total Number of normals: 317.
Somatic mutations of TRAM2-AS1:
Generating mutation plots.
Highly correlated genes for TRAM2-AS1:
Showing top 20/282 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRAM2-AS1 | TUBB2B | 0.698056 | 3 | 0 | 3 |
| TRAM2-AS1 | SGCE | 0.696835 | 6 | 0 | 6 |
| TRAM2-AS1 | ALDH1B1 | 0.692989 | 3 | 0 | 3 |
| TRAM2-AS1 | EML1 | 0.680338 | 3 | 0 | 3 |
| TRAM2-AS1 | TSPAN18 | 0.672513 | 4 | 0 | 3 |
| TRAM2-AS1 | MAGEH1 | 0.658016 | 5 | 0 | 5 |
| TRAM2-AS1 | KIAA1324 | 0.653982 | 3 | 0 | 3 |
| TRAM2-AS1 | TRAM2 | 0.651933 | 4 | 0 | 3 |
| TRAM2-AS1 | NEDD9 | 0.646383 | 5 | 0 | 4 |
| TRAM2-AS1 | F8 | 0.640288 | 5 | 0 | 4 |
| TRAM2-AS1 | ZNF275 | 0.639676 | 4 | 0 | 3 |
| TRAM2-AS1 | HMGN4 | 0.639309 | 4 | 0 | 3 |
| TRAM2-AS1 | CSGALNACT1 | 0.63915 | 4 | 0 | 4 |
| TRAM2-AS1 | LOH12CR2 | 0.636666 | 5 | 0 | 5 |
| TRAM2-AS1 | MAP9 | 0.635775 | 6 | 0 | 5 |
| TRAM2-AS1 | CLDN3 | 0.634916 | 3 | 0 | 3 |
| TRAM2-AS1 | KCNMA1 | 0.633356 | 4 | 0 | 4 |
| TRAM2-AS1 | LIPT1 | 0.632845 | 3 | 0 | 3 |
| TRAM2-AS1 | PIFO | 0.631335 | 3 | 0 | 3 |
| TRAM2-AS1 | BMP4 | 0.63082 | 5 | 0 | 4 |
For details and further investigation, click here