| Full name: thyrotropin releasing hormone degrading enzyme | Alias Symbol: PGPEP2|TRH-DE|PAP-II | ||
| Type: protein-coding gene | Cytoband: 12q21.1 | ||
| Entrez ID: 29953 | HGNC ID: HGNC:30748 | Ensembl Gene: ENSG00000072657 | OMIM ID: 606950 |
| Drug and gene relationship at DGIdb | |||
Expression of TRHDE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRHDE | 29953 | 219937_at | 0.7713 | 0.4383 | |
| GSE20347 | TRHDE | 29953 | 219937_at | 0.0589 | 0.5419 | |
| GSE23400 | TRHDE | 29953 | 219937_at | -0.0188 | 0.4166 | |
| GSE26886 | TRHDE | 29953 | 219937_at | -0.0701 | 0.5578 | |
| GSE29001 | TRHDE | 29953 | 219937_at | 0.3548 | 0.3394 | |
| GSE38129 | TRHDE | 29953 | 219937_at | -0.1811 | 0.2218 | |
| GSE45670 | TRHDE | 29953 | 219937_at | -0.7698 | 0.0015 | |
| GSE63941 | TRHDE | 29953 | 219937_at | -5.8293 | 0.0000 | |
| GSE77861 | TRHDE | 29953 | 219937_at | -0.0774 | 0.4092 | |
| SRP133303 | TRHDE | 29953 | RNAseq | -0.8117 | 0.3060 | |
| TCGA | TRHDE | 29953 | RNAseq | -1.8229 | 0.0162 |
Upregulated datasets: 0; Downregulated datasets: 2.
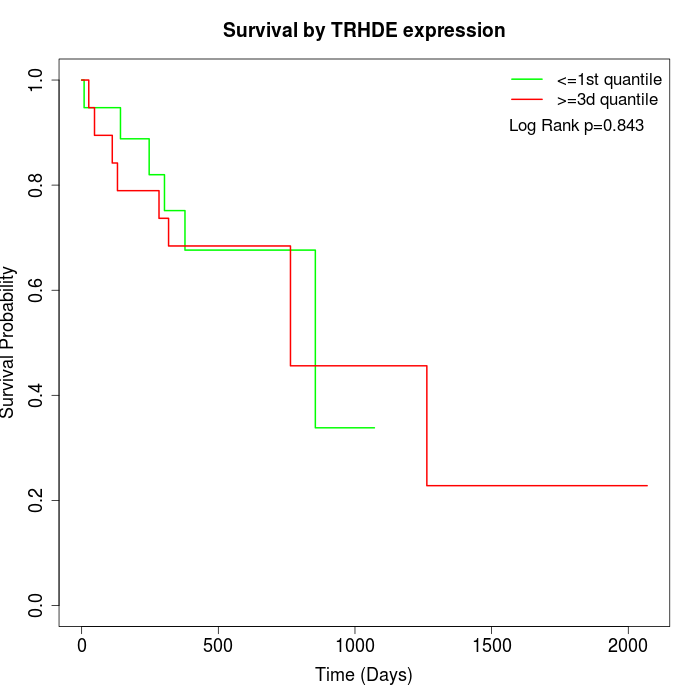
Survival by TRHDE expression:
Note: Click image to view full size file.
Copy number change of TRHDE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRHDE | 29953 | 4 | 3 | 23 | |
| GSE20123 | TRHDE | 29953 | 4 | 3 | 23 | |
| GSE43470 | TRHDE | 29953 | 3 | 0 | 40 | |
| GSE46452 | TRHDE | 29953 | 8 | 1 | 50 | |
| GSE47630 | TRHDE | 29953 | 10 | 1 | 29 | |
| GSE54993 | TRHDE | 29953 | 0 | 5 | 65 | |
| GSE54994 | TRHDE | 29953 | 5 | 2 | 46 | |
| GSE60625 | TRHDE | 29953 | 0 | 0 | 11 | |
| GSE74703 | TRHDE | 29953 | 3 | 0 | 33 | |
| GSE74704 | TRHDE | 29953 | 3 | 2 | 15 | |
| TCGA | TRHDE | 29953 | 19 | 10 | 67 |
Total number of gains: 59; Total number of losses: 27; Total Number of normals: 402.
Somatic mutations of TRHDE:
Generating mutation plots.
Highly correlated genes for TRHDE:
Showing top 20/62 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRHDE | AGTR1 | 0.73828 | 3 | 0 | 3 |
| TRHDE | DPP4 | 0.710647 | 3 | 0 | 3 |
| TRHDE | LAMA2 | 0.703611 | 3 | 0 | 3 |
| TRHDE | NOX4 | 0.696337 | 3 | 0 | 3 |
| TRHDE | ITGA1 | 0.692481 | 3 | 0 | 3 |
| TRHDE | CCDC102B | 0.681338 | 3 | 0 | 3 |
| TRHDE | THBS2 | 0.675241 | 3 | 0 | 3 |
| TRHDE | KLK14 | 0.669395 | 3 | 0 | 3 |
| TRHDE | DNM3 | 0.667818 | 4 | 0 | 3 |
| TRHDE | HAPLN1 | 0.657497 | 3 | 0 | 3 |
| TRHDE | MATN3 | 0.657465 | 3 | 0 | 3 |
| TRHDE | AFP | 0.654272 | 3 | 0 | 3 |
| TRHDE | DOCK4 | 0.653037 | 4 | 0 | 3 |
| TRHDE | INHBA | 0.653004 | 3 | 0 | 3 |
| TRHDE | OLFML1 | 0.64308 | 4 | 0 | 4 |
| TRHDE | SSC5D | 0.642243 | 3 | 0 | 3 |
| TRHDE | TRHDE-AS1 | 0.639946 | 4 | 0 | 3 |
| TRHDE | DBN1 | 0.639097 | 3 | 0 | 3 |
| TRHDE | TRIM8 | 0.631613 | 3 | 0 | 3 |
| TRHDE | CCL7 | 0.622686 | 3 | 0 | 3 |
For details and further investigation, click here