| Full name: tripartite motif containing 25 | Alias Symbol: EFP|RNF147 | ||
| Type: protein-coding gene | Cytoband: 17q23.1 | ||
| Entrez ID: 7706 | HGNC ID: HGNC:12932 | Ensembl Gene: ENSG00000121060 | OMIM ID: 600453 |
| Drug and gene relationship at DGIdb | |||
TRIM25 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04622 | RIG-I-like receptor signaling pathway | |
| hsa05164 | Influenza A |
Expression of TRIM25:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM25 | 7706 | 224806_at | 0.5916 | 0.1613 | |
| GSE20347 | TRIM25 | 7706 | 206911_at | -0.0000 | 0.9999 | |
| GSE23400 | TRIM25 | 7706 | 206911_at | -0.1112 | 0.0361 | |
| GSE26886 | TRIM25 | 7706 | 224806_at | -1.9837 | 0.0000 | |
| GSE29001 | TRIM25 | 7706 | 206911_at | -0.1821 | 0.2099 | |
| GSE38129 | TRIM25 | 7706 | 206911_at | -0.1039 | 0.1865 | |
| GSE45670 | TRIM25 | 7706 | 224806_at | 0.5757 | 0.0025 | |
| GSE53622 | TRIM25 | 7706 | 96801 | -0.4813 | 0.0000 | |
| GSE53624 | TRIM25 | 7706 | 96801 | -0.2269 | 0.0087 | |
| GSE63941 | TRIM25 | 7706 | 224806_at | 0.1839 | 0.7394 | |
| GSE77861 | TRIM25 | 7706 | 224806_at | 0.0719 | 0.7677 | |
| GSE97050 | TRIM25 | 7706 | A_33_P3331366 | -0.0910 | 0.7560 | |
| SRP007169 | TRIM25 | 7706 | RNAseq | -0.7566 | 0.0315 | |
| SRP008496 | TRIM25 | 7706 | RNAseq | -0.9011 | 0.0001 | |
| SRP064894 | TRIM25 | 7706 | RNAseq | -0.0565 | 0.7357 | |
| SRP133303 | TRIM25 | 7706 | RNAseq | -0.3583 | 0.0437 | |
| SRP159526 | TRIM25 | 7706 | RNAseq | -0.8684 | 0.0070 | |
| SRP193095 | TRIM25 | 7706 | RNAseq | -0.0639 | 0.6023 | |
| SRP219564 | TRIM25 | 7706 | RNAseq | 0.1000 | 0.8514 | |
| TCGA | TRIM25 | 7706 | RNAseq | -0.0513 | 0.3158 |
Upregulated datasets: 0; Downregulated datasets: 1.
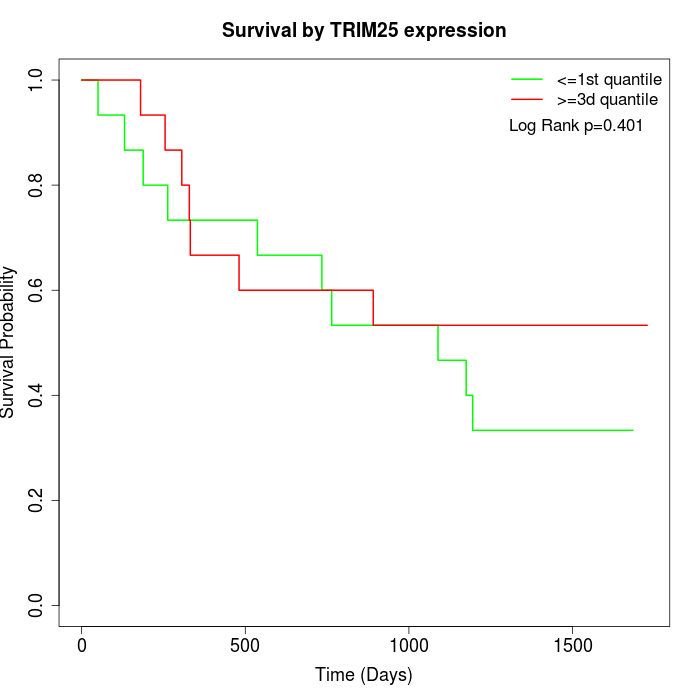
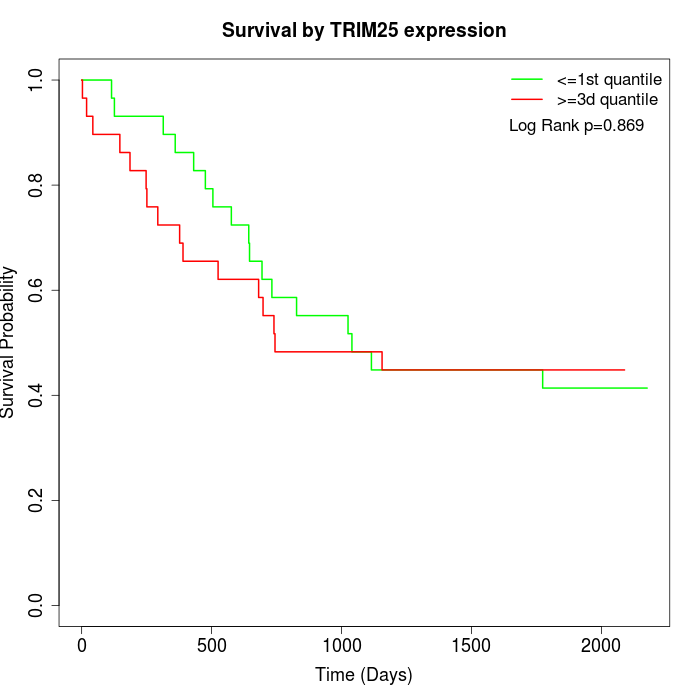
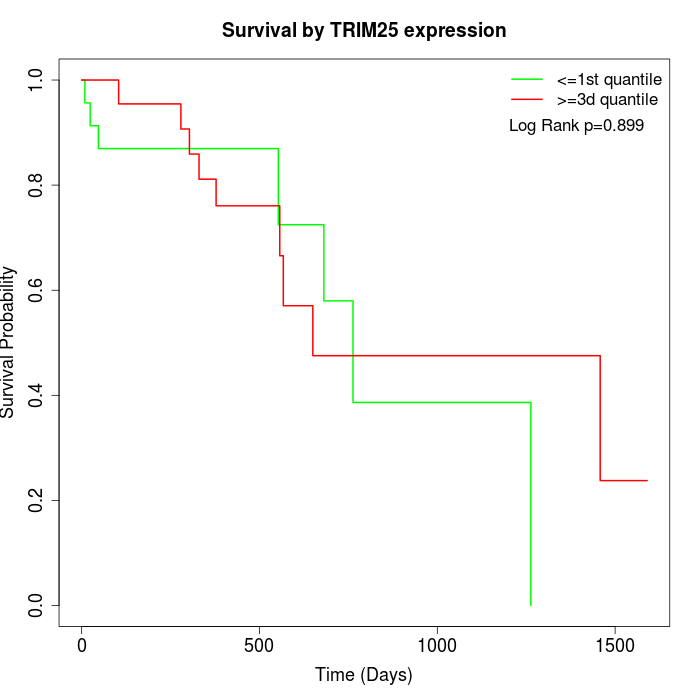
Survival by TRIM25 expression:
Note: Click image to view full size file.
Copy number change of TRIM25:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM25 | 7706 | 5 | 1 | 24 | |
| GSE20123 | TRIM25 | 7706 | 5 | 1 | 24 | |
| GSE43470 | TRIM25 | 7706 | 3 | 1 | 39 | |
| GSE46452 | TRIM25 | 7706 | 32 | 0 | 27 | |
| GSE47630 | TRIM25 | 7706 | 7 | 1 | 32 | |
| GSE54993 | TRIM25 | 7706 | 2 | 4 | 64 | |
| GSE54994 | TRIM25 | 7706 | 9 | 5 | 39 | |
| GSE60625 | TRIM25 | 7706 | 4 | 0 | 7 | |
| GSE74703 | TRIM25 | 7706 | 3 | 1 | 32 | |
| GSE74704 | TRIM25 | 7706 | 4 | 1 | 15 | |
| TCGA | TRIM25 | 7706 | 27 | 6 | 63 |
Total number of gains: 101; Total number of losses: 21; Total Number of normals: 366.
Somatic mutations of TRIM25:
Generating mutation plots.
Highly correlated genes for TRIM25:
Showing top 20/200 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM25 | STRADB | 0.700782 | 3 | 0 | 3 |
| TRIM25 | ULK3 | 0.685395 | 3 | 0 | 3 |
| TRIM25 | NAGK | 0.685112 | 3 | 0 | 3 |
| TRIM25 | GPT2 | 0.667175 | 3 | 0 | 3 |
| TRIM25 | PRPSAP1 | 0.654836 | 3 | 0 | 3 |
| TRIM25 | WSB2 | 0.638886 | 5 | 0 | 4 |
| TRIM25 | TOM1L2 | 0.63834 | 7 | 0 | 6 |
| TRIM25 | USP46 | 0.637581 | 4 | 0 | 3 |
| TRIM25 | KLRD1 | 0.635145 | 3 | 0 | 3 |
| TRIM25 | NAPA | 0.63405 | 4 | 0 | 3 |
| TRIM25 | C8A | 0.630991 | 4 | 0 | 3 |
| TRIM25 | CYB561D1 | 0.626801 | 4 | 0 | 4 |
| TRIM25 | CYP2D6 | 0.624611 | 4 | 0 | 4 |
| TRIM25 | IMPA2 | 0.623798 | 3 | 0 | 3 |
| TRIM25 | THEG | 0.622176 | 4 | 0 | 4 |
| TRIM25 | DRD2 | 0.61978 | 4 | 0 | 4 |
| TRIM25 | PLSCR1 | 0.616039 | 3 | 0 | 3 |
| TRIM25 | SNX11 | 0.614237 | 5 | 0 | 4 |
| TRIM25 | OPHN1 | 0.610715 | 5 | 0 | 4 |
| TRIM25 | ZNF92 | 0.608687 | 3 | 0 | 3 |
For details and further investigation, click here