| Full name: tripartite motif containing 28 | Alias Symbol: TIF1B|KAP1|TF1B|RNF96|PPP1R157 | ||
| Type: protein-coding gene | Cytoband: 19q13.4 | ||
| Entrez ID: 10155 | HGNC ID: HGNC:16384 | Ensembl Gene: ENSG00000130726 | OMIM ID: 601742 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRIM28:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM28 | 10155 | 200990_at | 0.8848 | 0.0652 | |
| GSE20347 | TRIM28 | 10155 | 200990_at | 0.4706 | 0.0029 | |
| GSE23400 | TRIM28 | 10155 | 200990_at | 0.5458 | 0.0000 | |
| GSE26886 | TRIM28 | 10155 | 200990_at | 0.5700 | 0.1052 | |
| GSE29001 | TRIM28 | 10155 | 200990_at | 0.4313 | 0.1841 | |
| GSE38129 | TRIM28 | 10155 | 200990_at | 0.5342 | 0.0000 | |
| GSE45670 | TRIM28 | 10155 | 200990_at | 0.5248 | 0.0033 | |
| GSE53622 | TRIM28 | 10155 | 93607 | 0.3251 | 0.0000 | |
| GSE53624 | TRIM28 | 10155 | 93607 | 0.6229 | 0.0000 | |
| GSE63941 | TRIM28 | 10155 | 200990_at | 2.4595 | 0.0000 | |
| GSE77861 | TRIM28 | 10155 | 200990_at | 0.5645 | 0.0818 | |
| GSE97050 | TRIM28 | 10155 | A_23_P89884 | 0.2903 | 0.2494 | |
| SRP007169 | TRIM28 | 10155 | RNAseq | -0.5871 | 0.2014 | |
| SRP008496 | TRIM28 | 10155 | RNAseq | -0.3082 | 0.2248 | |
| SRP064894 | TRIM28 | 10155 | RNAseq | 0.7299 | 0.0027 | |
| SRP133303 | TRIM28 | 10155 | RNAseq | 0.3262 | 0.1667 | |
| SRP159526 | TRIM28 | 10155 | RNAseq | 1.0402 | 0.0032 | |
| SRP193095 | TRIM28 | 10155 | RNAseq | 0.4746 | 0.0000 | |
| SRP219564 | TRIM28 | 10155 | RNAseq | 0.4763 | 0.1340 | |
| TCGA | TRIM28 | 10155 | RNAseq | 0.1213 | 0.0138 |
Upregulated datasets: 2; Downregulated datasets: 0.
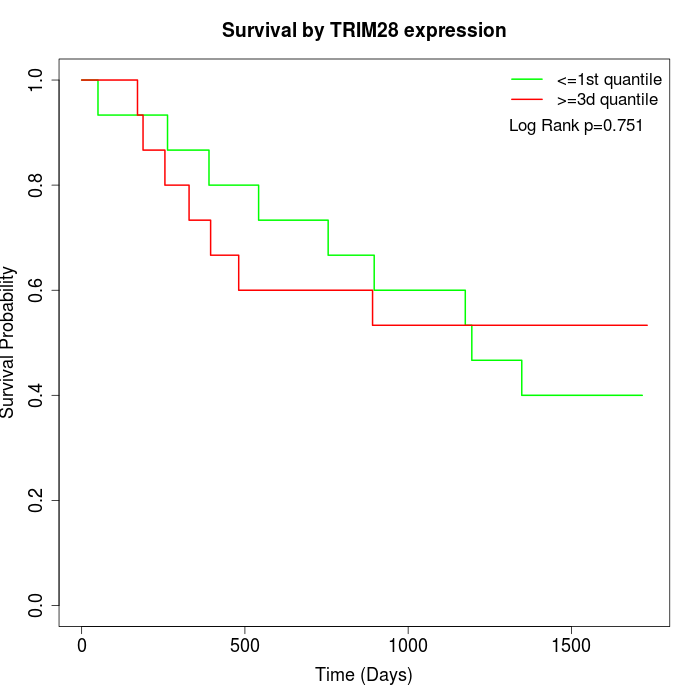
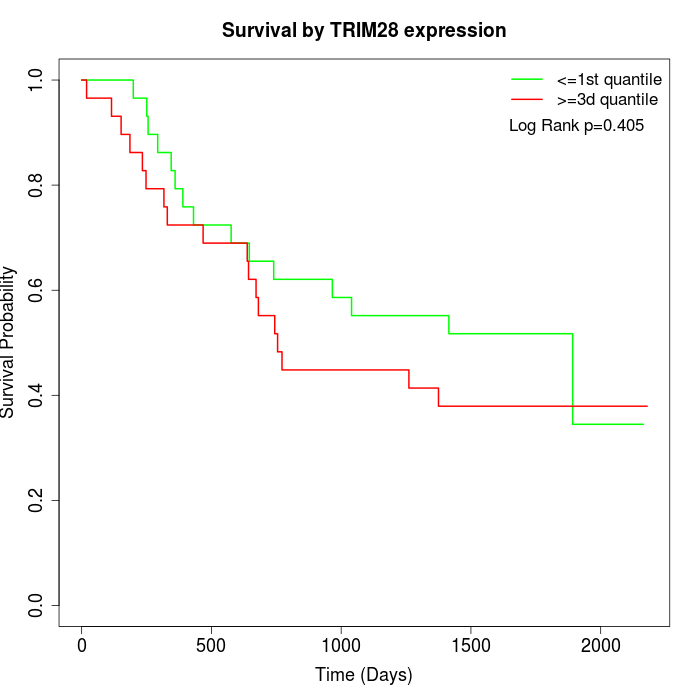
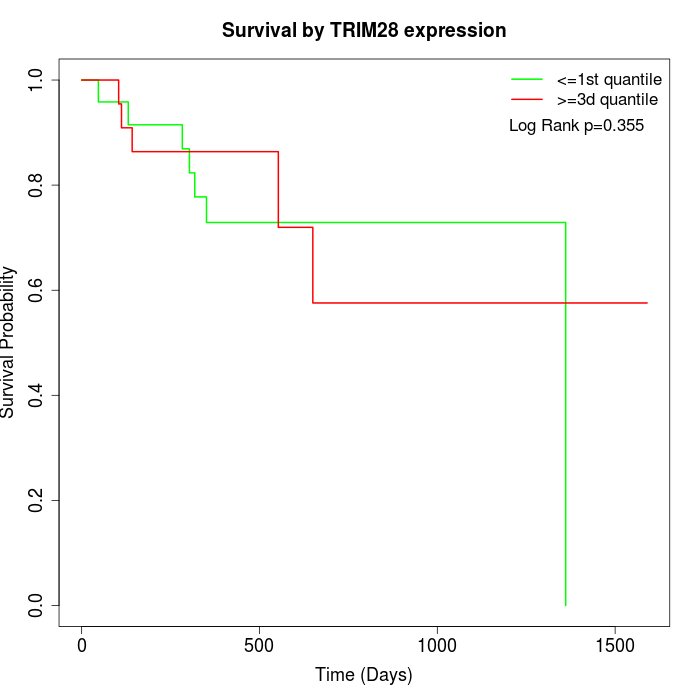
Survival by TRIM28 expression:
Note: Click image to view full size file.
Copy number change of TRIM28:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM28 | 10155 | 2 | 5 | 23 | |
| GSE20123 | TRIM28 | 10155 | 2 | 4 | 24 | |
| GSE43470 | TRIM28 | 10155 | 3 | 9 | 31 | |
| GSE46452 | TRIM28 | 10155 | 45 | 1 | 13 | |
| GSE47630 | TRIM28 | 10155 | 7 | 6 | 27 | |
| GSE54993 | TRIM28 | 10155 | 18 | 4 | 48 | |
| GSE54994 | TRIM28 | 10155 | 5 | 12 | 36 | |
| GSE60625 | TRIM28 | 10155 | 9 | 0 | 2 | |
| GSE74703 | TRIM28 | 10155 | 3 | 6 | 27 | |
| GSE74704 | TRIM28 | 10155 | 2 | 1 | 17 | |
| TCGA | TRIM28 | 10155 | 20 | 19 | 57 |
Total number of gains: 116; Total number of losses: 67; Total Number of normals: 305.
Somatic mutations of TRIM28:
Generating mutation plots.
Highly correlated genes for TRIM28:
Showing top 20/1502 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM28 | ADAM15 | 0.811898 | 3 | 0 | 3 |
| TRIM28 | LRRC41 | 0.799418 | 3 | 0 | 3 |
| TRIM28 | TRAPPC1 | 0.791773 | 3 | 0 | 3 |
| TRIM28 | NACC1 | 0.788724 | 4 | 0 | 3 |
| TRIM28 | TFDP2 | 0.772442 | 3 | 0 | 3 |
| TRIM28 | WDR89 | 0.771464 | 3 | 0 | 3 |
| TRIM28 | EIF3D | 0.769514 | 3 | 0 | 3 |
| TRIM28 | CBX6 | 0.747868 | 3 | 0 | 3 |
| TRIM28 | SNRPA | 0.737942 | 13 | 0 | 13 |
| TRIM28 | GORAB | 0.737577 | 4 | 0 | 4 |
| TRIM28 | VMA21 | 0.735346 | 3 | 0 | 3 |
| TRIM28 | TET1 | 0.733517 | 3 | 0 | 3 |
| TRIM28 | PGRMC1 | 0.733492 | 3 | 0 | 3 |
| TRIM28 | LRRC8B | 0.729172 | 3 | 0 | 3 |
| TRIM28 | ARFGAP2 | 0.725241 | 3 | 0 | 3 |
| TRIM28 | CHAMP1 | 0.724985 | 3 | 0 | 3 |
| TRIM28 | PRMT1 | 0.723745 | 11 | 0 | 10 |
| TRIM28 | KHSRP | 0.720584 | 8 | 0 | 8 |
| TRIM28 | SLC39A3 | 0.714677 | 4 | 0 | 4 |
| TRIM28 | PTBP1 | 0.713805 | 11 | 0 | 10 |
For details and further investigation, click here