| Full name: tripartite motif containing 61 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4q32.3 | ||
| Entrez ID: 391712 | HGNC ID: HGNC:24339 | Ensembl Gene: ENSG00000183439 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRIM61:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM61 | 391712 | 240342_at | -0.1066 | 0.6672 | |
| GSE26886 | TRIM61 | 391712 | 240342_at | -0.0210 | 0.8499 | |
| GSE45670 | TRIM61 | 391712 | 240342_at | -0.1528 | 0.0302 | |
| GSE53622 | TRIM61 | 391712 | 18008 | 0.2687 | 0.0014 | |
| GSE53624 | TRIM61 | 391712 | 18008 | 0.1771 | 0.0180 | |
| GSE63941 | TRIM61 | 391712 | 240342_at | -0.4846 | 0.0332 | |
| GSE77861 | TRIM61 | 391712 | 240342_at | -0.0668 | 0.5645 | |
| GSE97050 | TRIM61 | 391712 | A_33_P3383724 | 0.1699 | 0.5571 | |
| SRP133303 | TRIM61 | 391712 | RNAseq | -0.0820 | 0.6852 | |
| SRP159526 | TRIM61 | 391712 | RNAseq | -0.1665 | 0.7539 | |
| SRP193095 | TRIM61 | 391712 | RNAseq | -0.0219 | 0.8626 | |
| SRP219564 | TRIM61 | 391712 | RNAseq | 0.1834 | 0.6931 | |
| TCGA | TRIM61 | 391712 | RNAseq | -0.0042 | 0.9952 |
Upregulated datasets: 0; Downregulated datasets: 0.
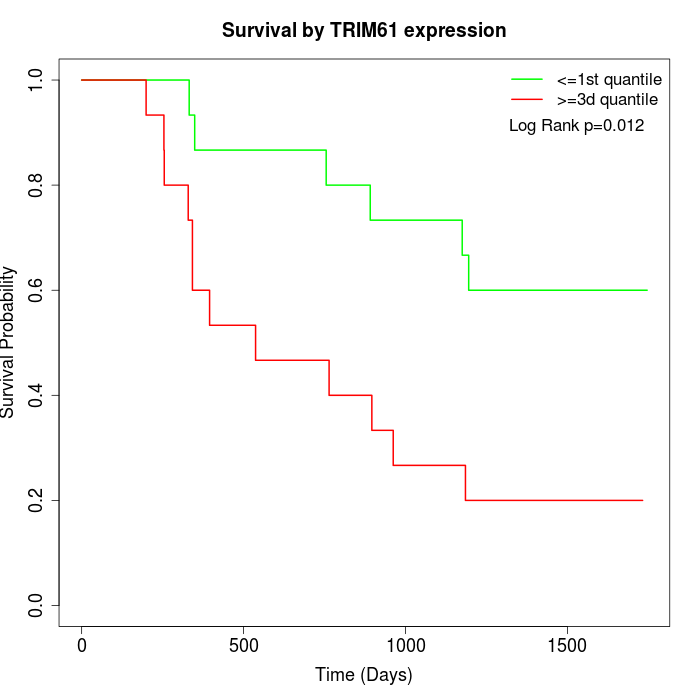
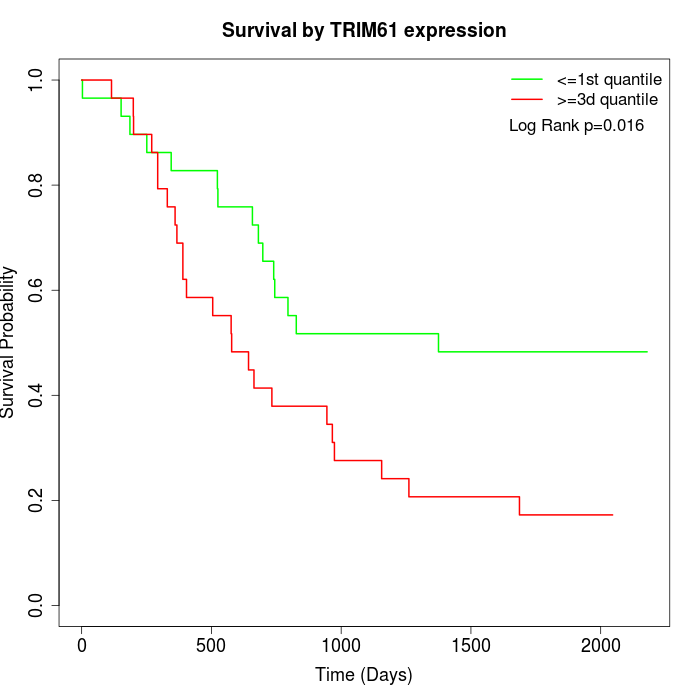
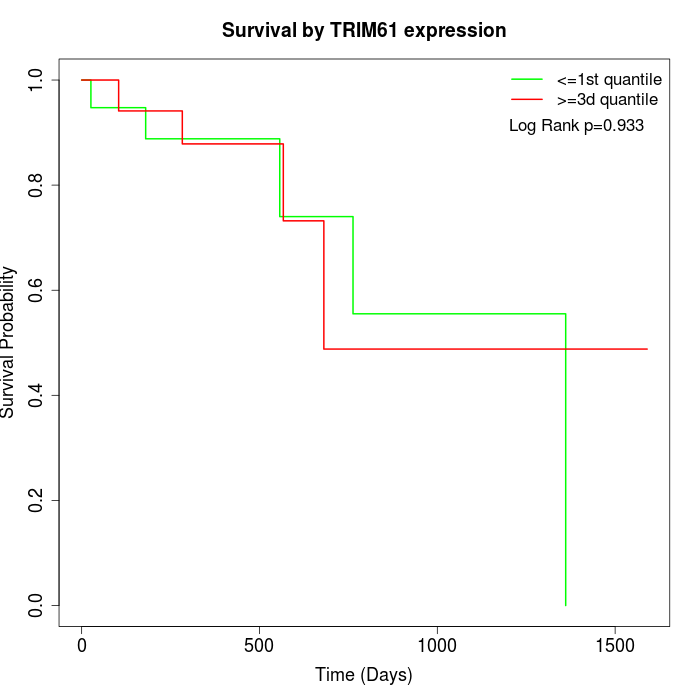
Survival by TRIM61 expression:
Note: Click image to view full size file.
Copy number change of TRIM61:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM61 | 391712 | 0 | 11 | 19 | |
| GSE20123 | TRIM61 | 391712 | 0 | 11 | 19 | |
| GSE43470 | TRIM61 | 391712 | 0 | 14 | 29 | |
| GSE46452 | TRIM61 | 391712 | 1 | 36 | 22 | |
| GSE47630 | TRIM61 | 391712 | 0 | 22 | 18 | |
| GSE54993 | TRIM61 | 391712 | 10 | 0 | 60 | |
| GSE54994 | TRIM61 | 391712 | 1 | 12 | 40 | |
| GSE60625 | TRIM61 | 391712 | 0 | 0 | 11 | |
| GSE74703 | TRIM61 | 391712 | 0 | 12 | 24 | |
| GSE74704 | TRIM61 | 391712 | 0 | 5 | 15 | |
| TCGA | TRIM61 | 391712 | 10 | 33 | 53 |
Total number of gains: 22; Total number of losses: 156; Total Number of normals: 310.
Somatic mutations of TRIM61:
Generating mutation plots.
Highly correlated genes for TRIM61:
Showing top 20/78 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM61 | SNCA | 0.824986 | 3 | 0 | 3 |
| TRIM61 | COMMD10 | 0.819744 | 3 | 0 | 3 |
| TRIM61 | BDKRB1 | 0.74188 | 3 | 0 | 3 |
| TRIM61 | C19orf47 | 0.740911 | 3 | 0 | 3 |
| TRIM61 | EIF3I | 0.736511 | 3 | 0 | 3 |
| TRIM61 | CHURC1 | 0.730849 | 3 | 0 | 3 |
| TRIM61 | SARDH | 0.729481 | 3 | 0 | 3 |
| TRIM61 | RBX1 | 0.727446 | 3 | 0 | 3 |
| TRIM61 | ANKRA2 | 0.720389 | 3 | 0 | 3 |
| TRIM61 | CCDC65 | 0.718556 | 3 | 0 | 3 |
| TRIM61 | LYRM7 | 0.715926 | 3 | 0 | 3 |
| TRIM61 | PAPPA | 0.711874 | 4 | 0 | 3 |
| TRIM61 | ADAMTSL5 | 0.706185 | 3 | 0 | 3 |
| TRIM61 | FYTTD1 | 0.695093 | 3 | 0 | 3 |
| TRIM61 | MTUS1 | 0.694721 | 3 | 0 | 3 |
| TRIM61 | MT4 | 0.694487 | 3 | 0 | 3 |
| TRIM61 | MRPS27 | 0.694009 | 4 | 0 | 3 |
| TRIM61 | UBE2J1 | 0.692905 | 3 | 0 | 3 |
| TRIM61 | NES | 0.683603 | 3 | 0 | 3 |
| TRIM61 | VPS29 | 0.683381 | 4 | 0 | 3 |
For details and further investigation, click here