| Full name: tripartite motif containing 65 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q25.1 | ||
| Entrez ID: 201292 | HGNC ID: HGNC:27316 | Ensembl Gene: ENSG00000141569 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM65:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM65 | 201292 | 235081_x_at | -0.0461 | 0.8819 | |
| GSE26886 | TRIM65 | 201292 | 235081_x_at | 0.0014 | 0.9877 | |
| GSE45670 | TRIM65 | 201292 | 235081_x_at | 0.1388 | 0.0793 | |
| GSE53622 | TRIM65 | 201292 | 109698 | 0.4415 | 0.0000 | |
| GSE53624 | TRIM65 | 201292 | 109698 | 0.6401 | 0.0000 | |
| GSE63941 | TRIM65 | 201292 | 235081_x_at | 0.7443 | 0.0002 | |
| GSE77861 | TRIM65 | 201292 | 235081_x_at | 0.0335 | 0.8666 | |
| GSE97050 | TRIM65 | 201292 | A_33_P3331346 | 0.3350 | 0.2124 | |
| SRP007169 | TRIM65 | 201292 | RNAseq | 0.9406 | 0.0295 | |
| SRP008496 | TRIM65 | 201292 | RNAseq | 0.9729 | 0.0012 | |
| SRP064894 | TRIM65 | 201292 | RNAseq | 0.6234 | 0.0000 | |
| SRP133303 | TRIM65 | 201292 | RNAseq | 0.5335 | 0.0034 | |
| SRP159526 | TRIM65 | 201292 | RNAseq | 0.4502 | 0.0335 | |
| SRP193095 | TRIM65 | 201292 | RNAseq | 0.3509 | 0.0002 | |
| SRP219564 | TRIM65 | 201292 | RNAseq | -0.0326 | 0.9026 | |
| TCGA | TRIM65 | 201292 | RNAseq | 0.2194 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 0.
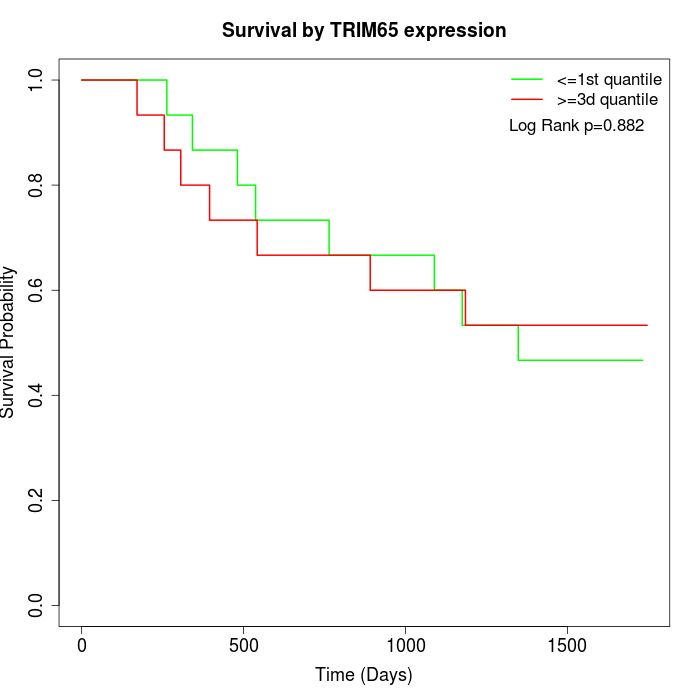
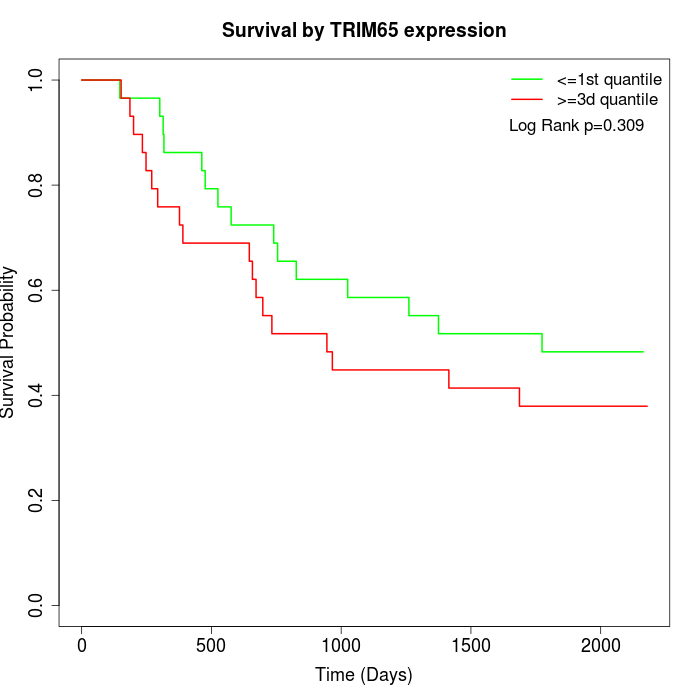
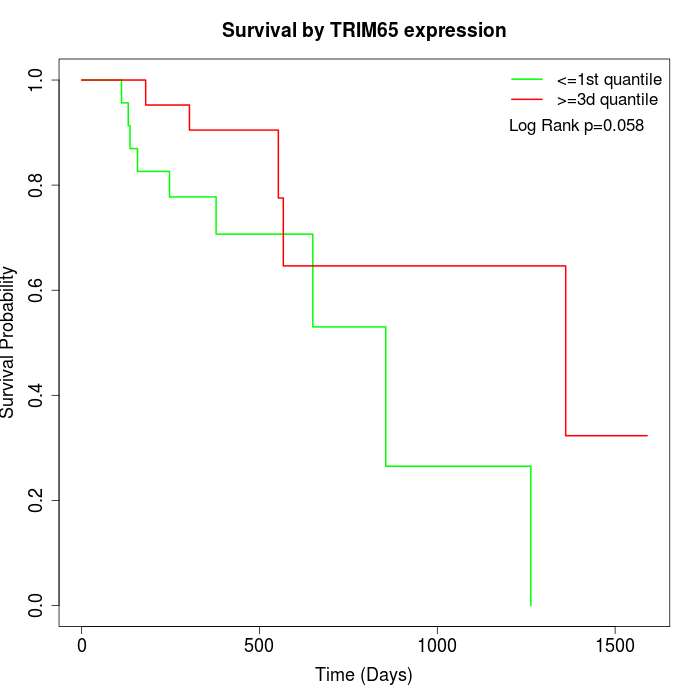
Survival by TRIM65 expression:
Note: Click image to view full size file.
Copy number change of TRIM65:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM65 | 201292 | 7 | 2 | 21 | |
| GSE20123 | TRIM65 | 201292 | 7 | 2 | 21 | |
| GSE43470 | TRIM65 | 201292 | 5 | 3 | 35 | |
| GSE46452 | TRIM65 | 201292 | 34 | 0 | 25 | |
| GSE47630 | TRIM65 | 201292 | 8 | 1 | 31 | |
| GSE54993 | TRIM65 | 201292 | 2 | 5 | 63 | |
| GSE54994 | TRIM65 | 201292 | 10 | 4 | 39 | |
| GSE60625 | TRIM65 | 201292 | 6 | 0 | 5 | |
| GSE74703 | TRIM65 | 201292 | 5 | 1 | 30 | |
| GSE74704 | TRIM65 | 201292 | 5 | 1 | 14 | |
| TCGA | TRIM65 | 201292 | 31 | 10 | 55 |
Total number of gains: 120; Total number of losses: 29; Total Number of normals: 339.
Somatic mutations of TRIM65:
Generating mutation plots.
Highly correlated genes for TRIM65:
Showing top 20/331 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM65 | EFTUD2 | 0.733883 | 4 | 0 | 4 |
| TRIM65 | ZNF207 | 0.732774 | 3 | 0 | 3 |
| TRIM65 | CBLC | 0.721317 | 3 | 0 | 3 |
| TRIM65 | CCDC65 | 0.718001 | 3 | 0 | 3 |
| TRIM65 | METTL23 | 0.71547 | 3 | 0 | 3 |
| TRIM65 | DDX39A | 0.708234 | 4 | 0 | 4 |
| TRIM65 | SLC16A5 | 0.706414 | 3 | 0 | 3 |
| TRIM65 | PLXNA1 | 0.702859 | 3 | 0 | 3 |
| TRIM65 | NCAPD2 | 0.698043 | 4 | 0 | 4 |
| TRIM65 | UBE2D2 | 0.694662 | 3 | 0 | 3 |
| TRIM65 | PTCD1 | 0.694616 | 3 | 0 | 3 |
| TRIM65 | MTHFD1L | 0.691257 | 3 | 0 | 3 |
| TRIM65 | GPR132 | 0.689456 | 3 | 0 | 3 |
| TRIM65 | ZNF707 | 0.685008 | 3 | 0 | 3 |
| TRIM65 | MTHFD1 | 0.684651 | 4 | 0 | 4 |
| TRIM65 | ZDHHC16 | 0.683887 | 3 | 0 | 3 |
| TRIM65 | TMEM138 | 0.68308 | 3 | 0 | 3 |
| TRIM65 | ATAD2 | 0.682742 | 4 | 0 | 4 |
| TRIM65 | CENPJ | 0.682319 | 3 | 0 | 3 |
| TRIM65 | POLE3 | 0.681745 | 5 | 0 | 4 |
For details and further investigation, click here