| Full name: tripartite motif containing 9 | Alias Symbol: SPRING|RNF91 | ||
| Type: protein-coding gene | Cytoband: 14q22.1 | ||
| Entrez ID: 114088 | HGNC ID: HGNC:16288 | Ensembl Gene: ENSG00000100505 | OMIM ID: 606555 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRIM9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM9 | 114088 | 209859_at | 1.0633 | 0.1411 | |
| GSE20347 | TRIM9 | 114088 | 209859_at | 0.7024 | 0.0037 | |
| GSE23400 | TRIM9 | 114088 | 209859_at | 0.2021 | 0.0002 | |
| GSE26886 | TRIM9 | 114088 | 209859_at | 1.8809 | 0.0001 | |
| GSE29001 | TRIM9 | 114088 | 209859_at | 0.9233 | 0.0070 | |
| GSE38129 | TRIM9 | 114088 | 209859_at | 0.5501 | 0.0047 | |
| GSE45670 | TRIM9 | 114088 | 209859_at | 0.2425 | 0.6026 | |
| GSE53622 | TRIM9 | 114088 | 66380 | 1.1326 | 0.0000 | |
| GSE53624 | TRIM9 | 114088 | 66380 | 1.5046 | 0.0000 | |
| GSE63941 | TRIM9 | 114088 | 209859_at | -0.0519 | 0.9618 | |
| GSE77861 | TRIM9 | 114088 | 209859_at | 0.2985 | 0.2339 | |
| GSE97050 | TRIM9 | 114088 | A_33_P3383836 | 0.5980 | 0.2827 | |
| SRP064894 | TRIM9 | 114088 | RNAseq | 1.6527 | 0.0001 | |
| SRP133303 | TRIM9 | 114088 | RNAseq | 0.5518 | 0.2138 | |
| SRP159526 | TRIM9 | 114088 | RNAseq | 1.7305 | 0.0322 | |
| SRP219564 | TRIM9 | 114088 | RNAseq | 1.2661 | 0.0140 | |
| TCGA | TRIM9 | 114088 | RNAseq | 0.0096 | 0.9724 |
Upregulated datasets: 6; Downregulated datasets: 0.
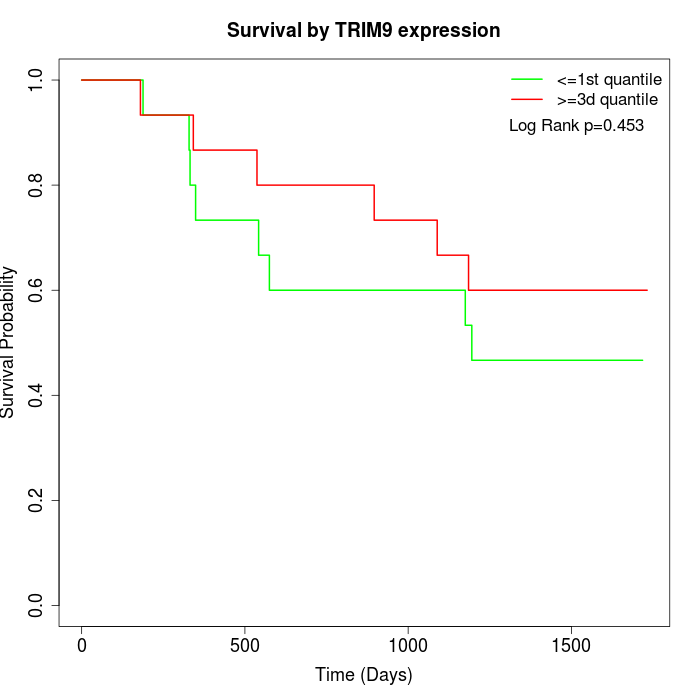
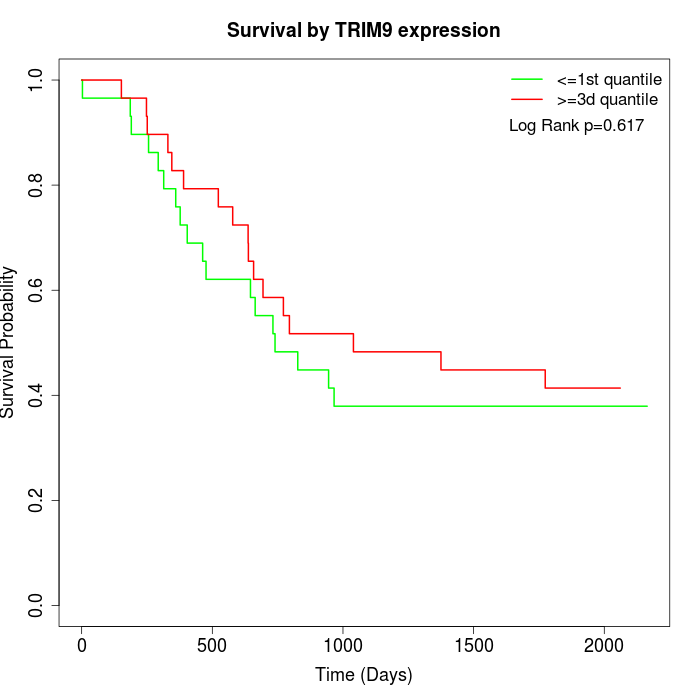
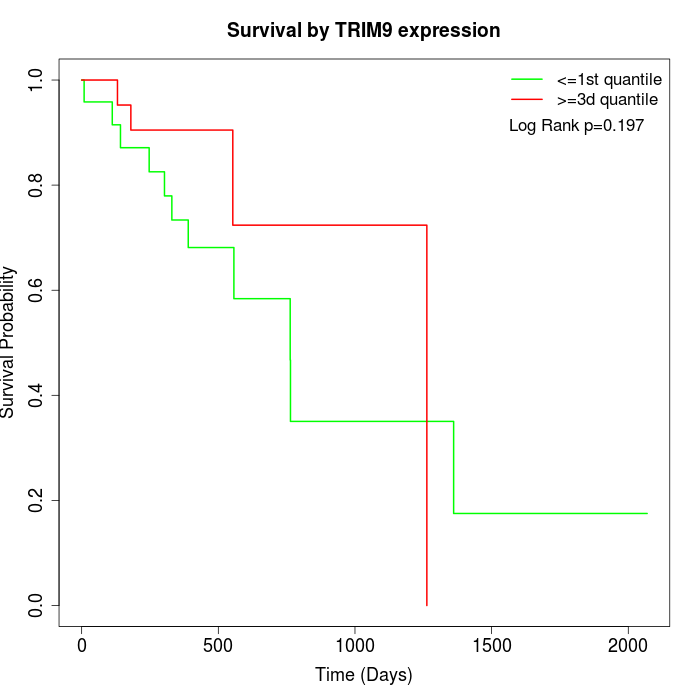
Survival by TRIM9 expression:
Note: Click image to view full size file.
Copy number change of TRIM9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM9 | 114088 | 8 | 3 | 19 | |
| GSE20123 | TRIM9 | 114088 | 8 | 3 | 19 | |
| GSE43470 | TRIM9 | 114088 | 9 | 1 | 33 | |
| GSE46452 | TRIM9 | 114088 | 16 | 3 | 40 | |
| GSE47630 | TRIM9 | 114088 | 11 | 10 | 19 | |
| GSE54993 | TRIM9 | 114088 | 3 | 8 | 59 | |
| GSE54994 | TRIM9 | 114088 | 19 | 4 | 30 | |
| GSE60625 | TRIM9 | 114088 | 0 | 2 | 9 | |
| GSE74703 | TRIM9 | 114088 | 8 | 1 | 27 | |
| GSE74704 | TRIM9 | 114088 | 3 | 2 | 15 | |
| TCGA | TRIM9 | 114088 | 34 | 13 | 49 |
Total number of gains: 119; Total number of losses: 50; Total Number of normals: 319.
Somatic mutations of TRIM9:
Generating mutation plots.
Highly correlated genes for TRIM9:
Showing top 20/348 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM9 | SLC52A1 | 0.753693 | 3 | 0 | 3 |
| TRIM9 | YY1 | 0.704008 | 3 | 0 | 3 |
| TRIM9 | ZNF284 | 0.686868 | 3 | 0 | 3 |
| TRIM9 | C1orf53 | 0.678881 | 3 | 0 | 3 |
| TRIM9 | ASCL2 | 0.673407 | 4 | 0 | 3 |
| TRIM9 | SS18L1 | 0.662744 | 4 | 0 | 4 |
| TRIM9 | GMEB2 | 0.654868 | 4 | 0 | 3 |
| TRIM9 | NACC1 | 0.653211 | 3 | 0 | 3 |
| TRIM9 | ASAP1-IT1 | 0.651694 | 3 | 0 | 3 |
| TRIM9 | ZNF579 | 0.650748 | 3 | 0 | 3 |
| TRIM9 | CCHCR1 | 0.648111 | 4 | 0 | 3 |
| TRIM9 | CBX2 | 0.647282 | 4 | 0 | 3 |
| TRIM9 | CEP131 | 0.644459 | 3 | 0 | 3 |
| TRIM9 | PLA2G15 | 0.643805 | 4 | 0 | 3 |
| TRIM9 | VANGL2 | 0.64054 | 3 | 0 | 3 |
| TRIM9 | F2R | 0.638729 | 5 | 0 | 4 |
| TRIM9 | KRTCAP2 | 0.638419 | 3 | 0 | 3 |
| TRIM9 | SEL1L | 0.63643 | 5 | 0 | 4 |
| TRIM9 | ANXA10 | 0.633888 | 4 | 0 | 3 |
| TRIM9 | TANGO2 | 0.630632 | 4 | 0 | 3 |
For details and further investigation, click here