| Full name: voltage dependent anion channel 1 | Alias Symbol: MGC111064|PORIN | ||
| Type: protein-coding gene | Cytoband: 5q31.1 | ||
| Entrez ID: 7416 | HGNC ID: HGNC:12669 | Ensembl Gene: ENSG00000213585 | OMIM ID: 604492 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
VDAC1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa05012 | Parkinson's disease |
Expression of VDAC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VDAC1 | 7416 | 212038_s_at | 0.0004 | 0.9994 | |
| GSE20347 | VDAC1 | 7416 | 212038_s_at | -0.5066 | 0.0070 | |
| GSE23400 | VDAC1 | 7416 | 212038_s_at | -0.0487 | 0.5973 | |
| GSE26886 | VDAC1 | 7416 | 212038_s_at | -1.9037 | 0.0000 | |
| GSE29001 | VDAC1 | 7416 | 212038_s_at | -0.3763 | 0.3680 | |
| GSE38129 | VDAC1 | 7416 | 212038_s_at | -0.0864 | 0.6591 | |
| GSE45670 | VDAC1 | 7416 | 212038_s_at | -0.0655 | 0.6806 | |
| GSE53622 | VDAC1 | 7416 | 120601 | -0.3708 | 0.0000 | |
| GSE53624 | VDAC1 | 7416 | 21285 | -0.2681 | 0.0000 | |
| GSE63941 | VDAC1 | 7416 | 212038_s_at | -0.2965 | 0.4723 | |
| GSE77861 | VDAC1 | 7416 | 212038_s_at | -0.2934 | 0.3280 | |
| GSE97050 | VDAC1 | 7416 | A_23_P144816 | -0.0059 | 0.9830 | |
| SRP007169 | VDAC1 | 7416 | RNAseq | -1.5962 | 0.0000 | |
| SRP008496 | VDAC1 | 7416 | RNAseq | -1.4010 | 0.0000 | |
| SRP064894 | VDAC1 | 7416 | RNAseq | -0.4059 | 0.0022 | |
| SRP133303 | VDAC1 | 7416 | RNAseq | 0.1095 | 0.5820 | |
| SRP159526 | VDAC1 | 7416 | RNAseq | -0.3246 | 0.3444 | |
| SRP193095 | VDAC1 | 7416 | RNAseq | -0.6572 | 0.0000 | |
| SRP219564 | VDAC1 | 7416 | RNAseq | -0.2988 | 0.4761 | |
| TCGA | VDAC1 | 7416 | RNAseq | -0.0255 | 0.5670 |
Upregulated datasets: 0; Downregulated datasets: 3.
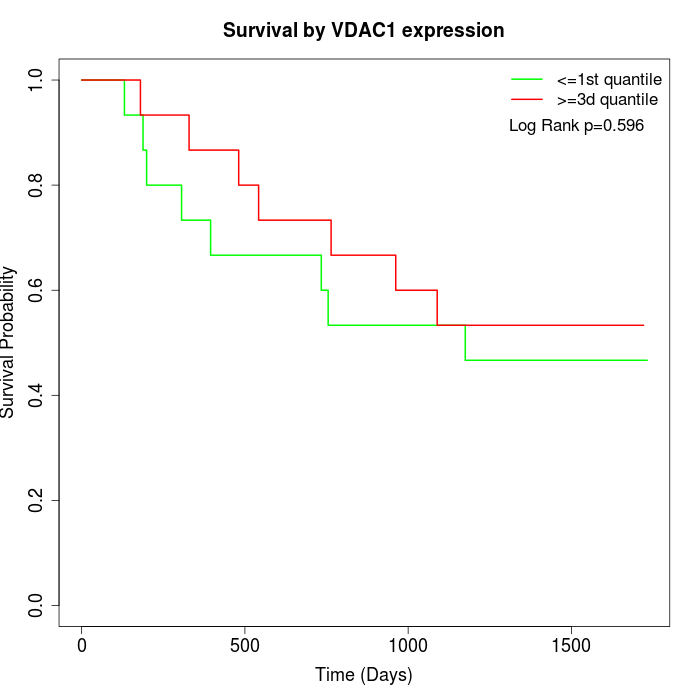
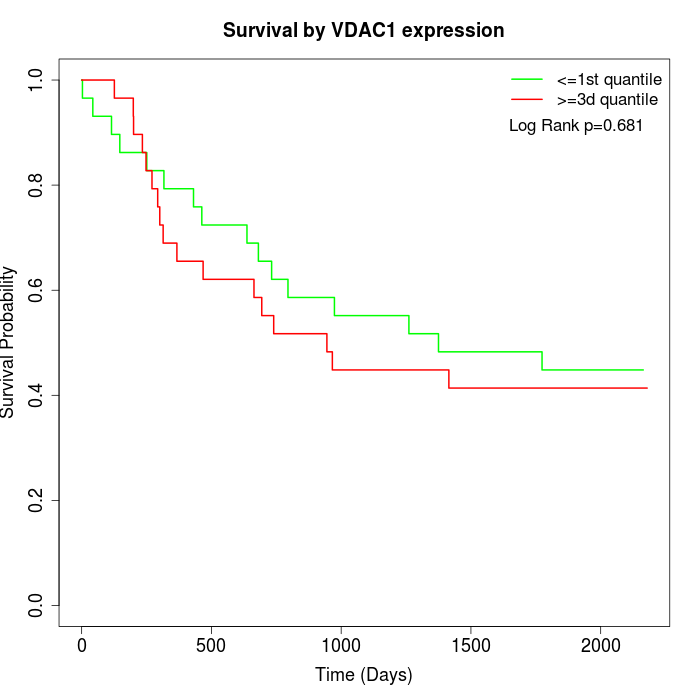
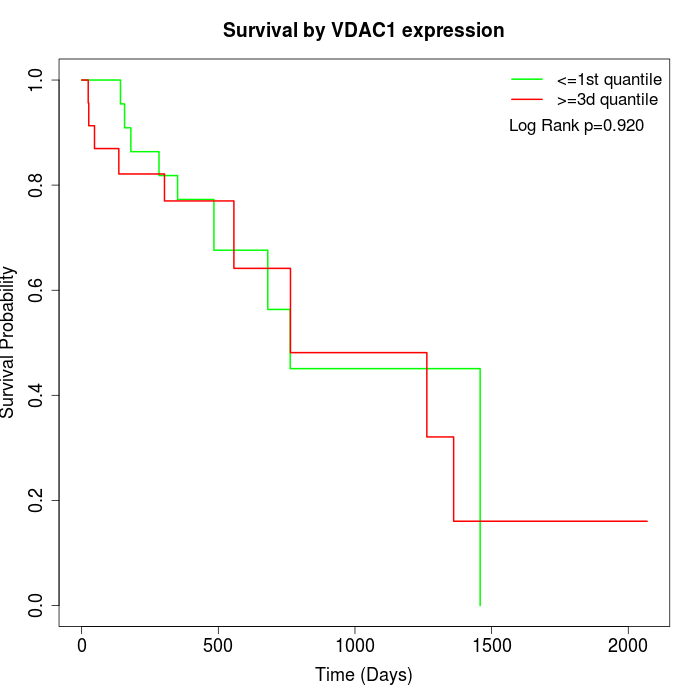
Survival by VDAC1 expression:
Note: Click image to view full size file.
Copy number change of VDAC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VDAC1 | 7416 | 2 | 11 | 17 | |
| GSE20123 | VDAC1 | 7416 | 3 | 11 | 16 | |
| GSE43470 | VDAC1 | 7416 | 2 | 9 | 32 | |
| GSE46452 | VDAC1 | 7416 | 0 | 27 | 32 | |
| GSE47630 | VDAC1 | 7416 | 0 | 21 | 19 | |
| GSE54993 | VDAC1 | 7416 | 9 | 1 | 60 | |
| GSE54994 | VDAC1 | 7416 | 2 | 14 | 37 | |
| GSE60625 | VDAC1 | 7416 | 0 | 0 | 11 | |
| GSE74703 | VDAC1 | 7416 | 2 | 6 | 28 | |
| GSE74704 | VDAC1 | 7416 | 2 | 5 | 13 | |
| TCGA | VDAC1 | 7416 | 3 | 37 | 56 |
Total number of gains: 25; Total number of losses: 142; Total Number of normals: 321.
Somatic mutations of VDAC1:
Generating mutation plots.
Highly correlated genes for VDAC1:
Showing top 20/695 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VDAC1 | ZMAT2 | 0.731748 | 5 | 0 | 5 |
| VDAC1 | PPP2CA | 0.731589 | 12 | 0 | 10 |
| VDAC1 | BNIPL | 0.724694 | 5 | 0 | 5 |
| VDAC1 | EMP2 | 0.692116 | 3 | 0 | 3 |
| VDAC1 | ABLIM2 | 0.689747 | 4 | 0 | 4 |
| VDAC1 | SLC35A4 | 0.684701 | 5 | 0 | 5 |
| VDAC1 | PLEKHM1 | 0.674676 | 4 | 0 | 4 |
| VDAC1 | HECTD1 | 0.670303 | 4 | 0 | 3 |
| VDAC1 | DIAPH1 | 0.66949 | 9 | 0 | 8 |
| VDAC1 | ARL2BP | 0.668853 | 4 | 0 | 4 |
| VDAC1 | TMEM154 | 0.663798 | 5 | 0 | 5 |
| VDAC1 | HSPA4 | 0.663642 | 12 | 0 | 9 |
| VDAC1 | GSKIP | 0.660691 | 5 | 0 | 5 |
| VDAC1 | SH3RF2 | 0.657874 | 4 | 0 | 4 |
| VDAC1 | PRDX5 | 0.656956 | 6 | 0 | 5 |
| VDAC1 | UBAC2 | 0.656539 | 4 | 0 | 4 |
| VDAC1 | RNPEP | 0.654926 | 7 | 0 | 6 |
| VDAC1 | RHOA | 0.654561 | 7 | 0 | 5 |
| VDAC1 | SMIM15 | 0.653921 | 4 | 0 | 4 |
| VDAC1 | SDR9C7 | 0.650513 | 5 | 0 | 4 |
For details and further investigation, click here