| Full name: diaphanous related formin 1 | Alias Symbol: hDIA1|LFHL1 | ||
| Type: protein-coding gene | Cytoband: 5q31.3 | ||
| Entrez ID: 1729 | HGNC ID: HGNC:2876 | Ensembl Gene: ENSG00000131504 | OMIM ID: 602121 |
| Drug and gene relationship at DGIdb | |||
DIAPH1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04510 | Focal adhesion | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications |
Expression of DIAPH1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DIAPH1 | 1729 | 209190_s_at | -0.0137 | 0.9895 | |
| GSE20347 | DIAPH1 | 1729 | 209190_s_at | -0.4616 | 0.0027 | |
| GSE23400 | DIAPH1 | 1729 | 209190_s_at | -0.3177 | 0.0006 | |
| GSE26886 | DIAPH1 | 1729 | 209190_s_at | -1.2156 | 0.0000 | |
| GSE29001 | DIAPH1 | 1729 | 209190_s_at | -0.4583 | 0.0109 | |
| GSE38129 | DIAPH1 | 1729 | 209190_s_at | -0.1656 | 0.4200 | |
| GSE45670 | DIAPH1 | 1729 | 209190_s_at | -0.1367 | 0.3497 | |
| GSE53622 | DIAPH1 | 1729 | 96767 | -0.4053 | 0.0004 | |
| GSE53624 | DIAPH1 | 1729 | 29679 | -0.6091 | 0.0000 | |
| GSE63941 | DIAPH1 | 1729 | 209190_s_at | 1.0636 | 0.0017 | |
| GSE77861 | DIAPH1 | 1729 | 209190_s_at | -0.4846 | 0.1283 | |
| GSE97050 | DIAPH1 | 1729 | A_33_P3243683 | 0.4705 | 0.4226 | |
| SRP007169 | DIAPH1 | 1729 | RNAseq | -1.4957 | 0.0008 | |
| SRP008496 | DIAPH1 | 1729 | RNAseq | -1.7056 | 0.0000 | |
| SRP064894 | DIAPH1 | 1729 | RNAseq | -0.9184 | 0.0000 | |
| SRP133303 | DIAPH1 | 1729 | RNAseq | -0.2047 | 0.3122 | |
| SRP159526 | DIAPH1 | 1729 | RNAseq | -0.9466 | 0.0133 | |
| SRP193095 | DIAPH1 | 1729 | RNAseq | -0.5681 | 0.0001 | |
| SRP219564 | DIAPH1 | 1729 | RNAseq | -0.5741 | 0.3620 | |
| TCGA | DIAPH1 | 1729 | RNAseq | -0.0255 | 0.6046 |
Upregulated datasets: 1; Downregulated datasets: 3.
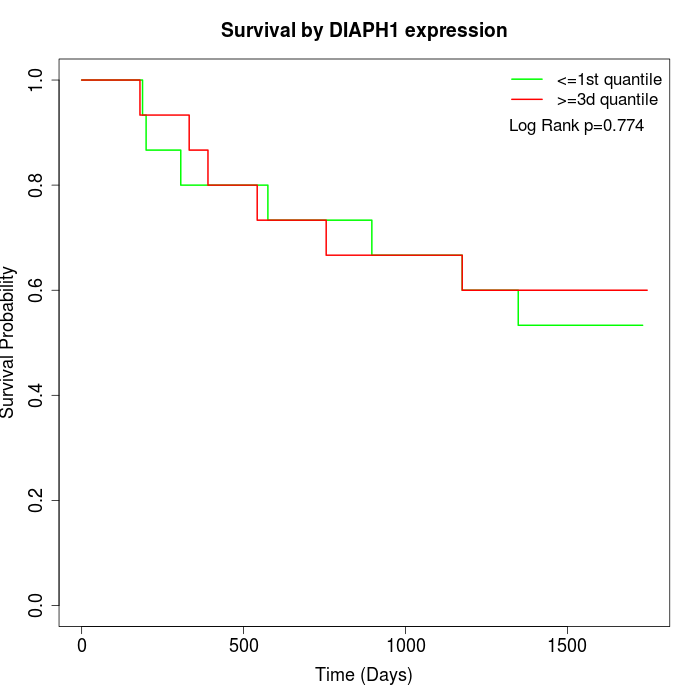
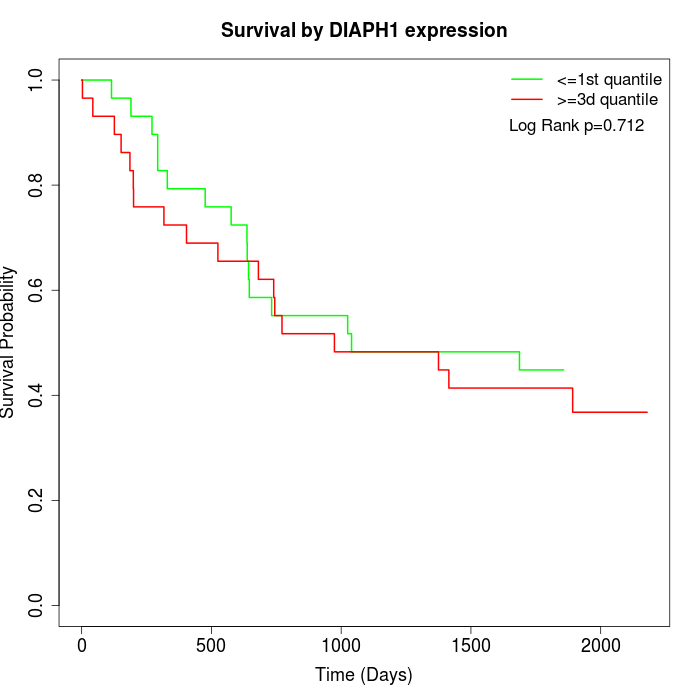
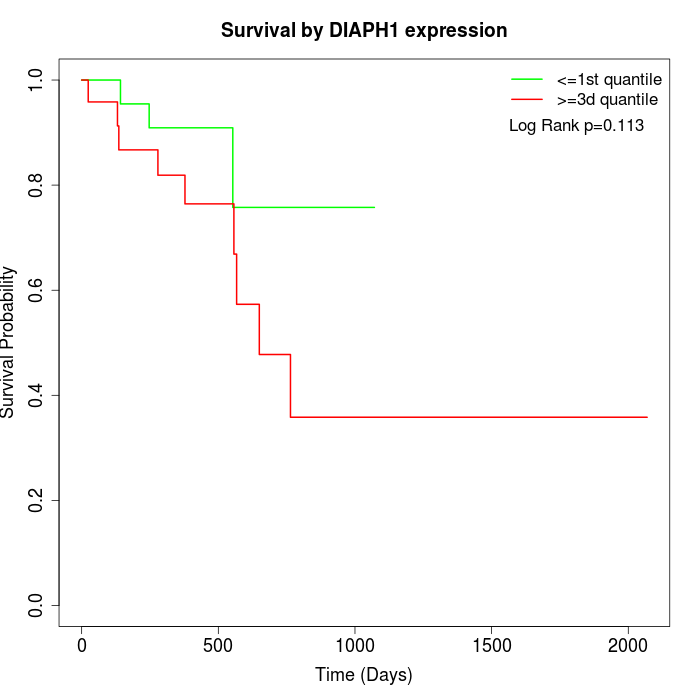
Survival by DIAPH1 expression:
Note: Click image to view full size file.
Copy number change of DIAPH1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DIAPH1 | 1729 | 2 | 11 | 17 | |
| GSE20123 | DIAPH1 | 1729 | 3 | 11 | 16 | |
| GSE43470 | DIAPH1 | 1729 | 2 | 10 | 31 | |
| GSE46452 | DIAPH1 | 1729 | 0 | 27 | 32 | |
| GSE47630 | DIAPH1 | 1729 | 1 | 19 | 20 | |
| GSE54993 | DIAPH1 | 1729 | 9 | 1 | 60 | |
| GSE54994 | DIAPH1 | 1729 | 2 | 14 | 37 | |
| GSE60625 | DIAPH1 | 1729 | 0 | 0 | 11 | |
| GSE74703 | DIAPH1 | 1729 | 2 | 7 | 27 | |
| GSE74704 | DIAPH1 | 1729 | 2 | 5 | 13 | |
| TCGA | DIAPH1 | 1729 | 3 | 38 | 55 |
Total number of gains: 26; Total number of losses: 143; Total Number of normals: 319.
Somatic mutations of DIAPH1:
Generating mutation plots.
Highly correlated genes for DIAPH1:
Showing top 20/1167 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DIAPH1 | KCNK6 | 0.799676 | 5 | 0 | 5 |
| DIAPH1 | AP1S3 | 0.781277 | 4 | 0 | 4 |
| DIAPH1 | CAPNS2 | 0.776877 | 3 | 0 | 3 |
| DIAPH1 | RIPK3 | 0.757589 | 5 | 0 | 5 |
| DIAPH1 | FAM122A | 0.748598 | 3 | 0 | 3 |
| DIAPH1 | RABGGTA | 0.746017 | 11 | 0 | 11 |
| DIAPH1 | FASN | 0.731976 | 3 | 0 | 3 |
| DIAPH1 | TOM1 | 0.730903 | 11 | 0 | 11 |
| DIAPH1 | ACTR2 | 0.71427 | 4 | 0 | 4 |
| DIAPH1 | DDI2 | 0.713393 | 3 | 0 | 3 |
| DIAPH1 | NUAK2 | 0.710422 | 3 | 0 | 3 |
| DIAPH1 | NIPAL1 | 0.709992 | 3 | 0 | 3 |
| DIAPH1 | DEF8 | 0.709452 | 4 | 0 | 4 |
| DIAPH1 | WDR77 | 0.708344 | 3 | 0 | 3 |
| DIAPH1 | PLIN3 | 0.708218 | 11 | 0 | 11 |
| DIAPH1 | RAB8A | 0.707809 | 4 | 0 | 3 |
| DIAPH1 | TMEM106A | 0.706998 | 3 | 0 | 3 |
| DIAPH1 | IQGAP3 | 0.706818 | 3 | 0 | 3 |
| DIAPH1 | GALE | 0.700795 | 13 | 0 | 12 |
| DIAPH1 | FAM83B | 0.700275 | 4 | 0 | 3 |
For details and further investigation, click here