| Full name: voltage dependent anion channel 3 | Alias Symbol: HD-VDAC3 | ||
| Type: protein-coding gene | Cytoband: 8p11.21 | ||
| Entrez ID: 7419 | HGNC ID: HGNC:12674 | Ensembl Gene: ENSG00000078668 | OMIM ID: 610029 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
VDAC3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa05012 | Parkinson's disease | |
| hsa05161 | Hepatitis B |
Expression of VDAC3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VDAC3 | 7419 | 208845_at | -0.2050 | 0.4990 | |
| GSE20347 | VDAC3 | 7419 | 208845_at | 0.0885 | 0.4560 | |
| GSE23400 | VDAC3 | 7419 | 208845_at | -0.0568 | 0.5204 | |
| GSE26886 | VDAC3 | 7419 | 208845_at | -0.3226 | 0.0070 | |
| GSE29001 | VDAC3 | 7419 | 208845_at | -0.0322 | 0.8566 | |
| GSE38129 | VDAC3 | 7419 | 208845_at | -0.0715 | 0.5912 | |
| GSE45670 | VDAC3 | 7419 | 208845_at | 0.0331 | 0.8410 | |
| GSE53622 | VDAC3 | 7419 | 2782 | -0.0903 | 0.2113 | |
| GSE53624 | VDAC3 | 7419 | 2782 | -0.1472 | 0.0393 | |
| GSE63941 | VDAC3 | 7419 | 208845_at | -0.0158 | 0.9733 | |
| GSE77861 | VDAC3 | 7419 | 208845_at | -0.2747 | 0.4027 | |
| GSE97050 | VDAC3 | 7419 | A_24_P235316 | 0.0526 | 0.8151 | |
| SRP007169 | VDAC3 | 7419 | RNAseq | -0.1040 | 0.7830 | |
| SRP008496 | VDAC3 | 7419 | RNAseq | -0.1904 | 0.4300 | |
| SRP064894 | VDAC3 | 7419 | RNAseq | -0.2001 | 0.2072 | |
| SRP133303 | VDAC3 | 7419 | RNAseq | 0.1063 | 0.5923 | |
| SRP159526 | VDAC3 | 7419 | RNAseq | -0.1818 | 0.5382 | |
| SRP193095 | VDAC3 | 7419 | RNAseq | -0.3783 | 0.0091 | |
| SRP219564 | VDAC3 | 7419 | RNAseq | -0.6781 | 0.0566 | |
| TCGA | VDAC3 | 7419 | RNAseq | 0.1352 | 0.0772 |
Upregulated datasets: 0; Downregulated datasets: 0.
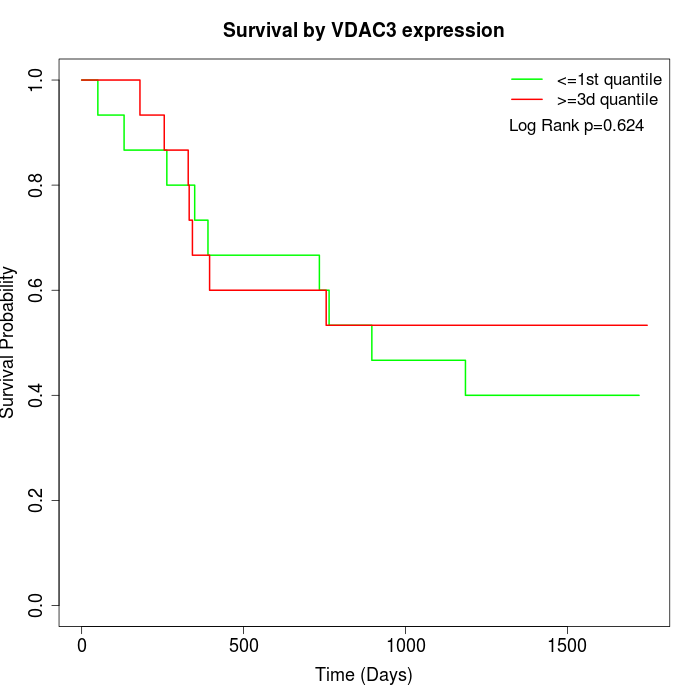
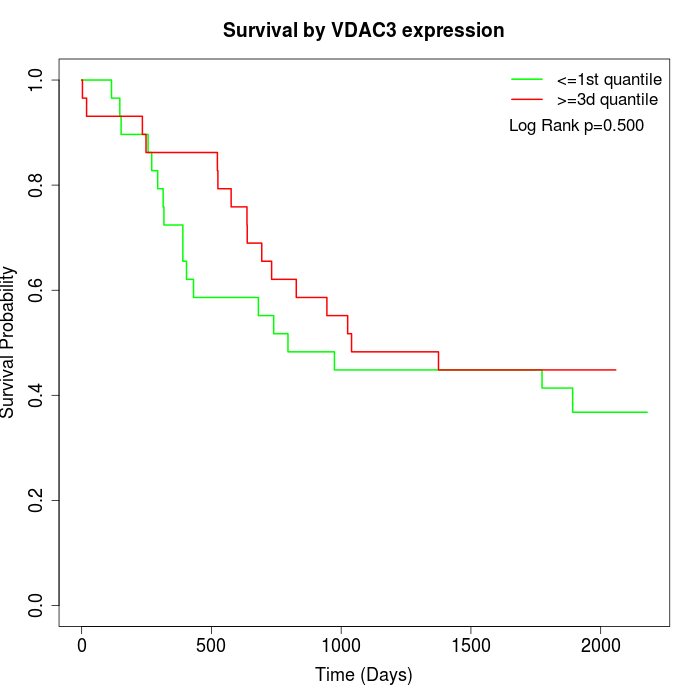
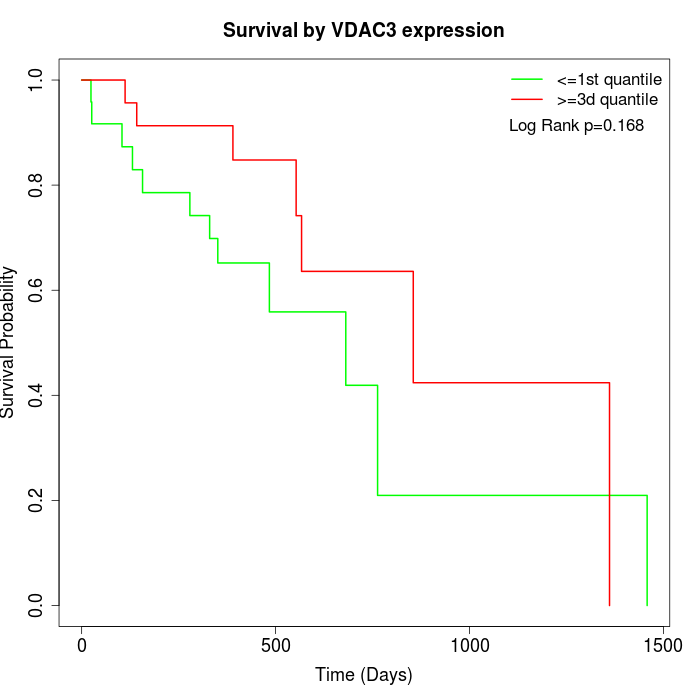
Survival by VDAC3 expression:
Note: Click image to view full size file.
Copy number change of VDAC3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VDAC3 | 7419 | 10 | 5 | 15 | |
| GSE20123 | VDAC3 | 7419 | 10 | 5 | 15 | |
| GSE43470 | VDAC3 | 7419 | 9 | 6 | 28 | |
| GSE46452 | VDAC3 | 7419 | 19 | 5 | 35 | |
| GSE47630 | VDAC3 | 7419 | 22 | 1 | 17 | |
| GSE54993 | VDAC3 | 7419 | 1 | 16 | 53 | |
| GSE54994 | VDAC3 | 7419 | 19 | 7 | 27 | |
| GSE60625 | VDAC3 | 7419 | 3 | 0 | 8 | |
| GSE74703 | VDAC3 | 7419 | 8 | 5 | 23 | |
| GSE74704 | VDAC3 | 7419 | 9 | 2 | 9 | |
| TCGA | VDAC3 | 7419 | 41 | 21 | 34 |
Total number of gains: 151; Total number of losses: 73; Total Number of normals: 264.
Somatic mutations of VDAC3:
Generating mutation plots.
Highly correlated genes for VDAC3:
Showing top 20/261 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VDAC3 | SRP14 | 0.842257 | 3 | 0 | 3 |
| VDAC3 | ARIH1 | 0.794924 | 3 | 0 | 3 |
| VDAC3 | UFL1 | 0.763214 | 3 | 0 | 3 |
| VDAC3 | TPRG1L | 0.762085 | 3 | 0 | 3 |
| VDAC3 | GLUD2 | 0.760473 | 3 | 0 | 3 |
| VDAC3 | PCYOX1 | 0.75729 | 3 | 0 | 3 |
| VDAC3 | RBKS | 0.75357 | 3 | 0 | 3 |
| VDAC3 | NDUFS1 | 0.749255 | 3 | 0 | 3 |
| VDAC3 | SOX7 | 0.748957 | 3 | 0 | 3 |
| VDAC3 | TMEM167B | 0.748783 | 3 | 0 | 3 |
| VDAC3 | PSMD8 | 0.743882 | 3 | 0 | 3 |
| VDAC3 | CHMP2A | 0.73845 | 3 | 0 | 3 |
| VDAC3 | AQR | 0.733412 | 4 | 0 | 4 |
| VDAC3 | RAB2A | 0.732176 | 3 | 0 | 3 |
| VDAC3 | RNF185 | 0.730975 | 3 | 0 | 3 |
| VDAC3 | EEA1 | 0.729422 | 3 | 0 | 3 |
| VDAC3 | CCNDBP1 | 0.725852 | 3 | 0 | 3 |
| VDAC3 | RNF141 | 0.721957 | 3 | 0 | 3 |
| VDAC3 | PTPRG | 0.720327 | 3 | 0 | 3 |
| VDAC3 | PKP2 | 0.719645 | 3 | 0 | 3 |
For details and further investigation, click here