| Full name: vimentin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10p13 | ||
| Entrez ID: 7431 | HGNC ID: HGNC:12692 | Ensembl Gene: ENSG00000026025 | OMIM ID: 193060 |
| Drug and gene relationship at DGIdb | |||
Expression of VIM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VIM | 7431 | 201426_s_at | -0.3753 | 0.7211 | |
| GSE20347 | VIM | 7431 | 201426_s_at | 0.7564 | 0.0289 | |
| GSE23400 | VIM | 7431 | 201426_s_at | 0.2884 | 0.1118 | |
| GSE26886 | VIM | 7431 | 201426_s_at | 1.3189 | 0.0019 | |
| GSE29001 | VIM | 7431 | 201426_s_at | 0.7619 | 0.2171 | |
| GSE38129 | VIM | 7431 | 201426_s_at | 0.3032 | 0.5673 | |
| GSE45670 | VIM | 7431 | 201426_s_at | -1.4511 | 0.0040 | |
| GSE53622 | VIM | 7431 | 45368 | 0.4248 | 0.0002 | |
| GSE53624 | VIM | 7431 | 45368 | 0.4845 | 0.0000 | |
| GSE63941 | VIM | 7431 | 201426_s_at | -6.0186 | 0.0037 | |
| GSE77861 | VIM | 7431 | 201426_s_at | 1.3756 | 0.0761 | |
| GSE97050 | VIM | 7431 | A_23_P161190 | 0.6483 | 0.1880 | |
| SRP007169 | VIM | 7431 | RNAseq | 2.2603 | 0.0001 | |
| SRP008496 | VIM | 7431 | RNAseq | 2.1075 | 0.0000 | |
| SRP064894 | VIM | 7431 | RNAseq | 0.6213 | 0.0676 | |
| SRP133303 | VIM | 7431 | RNAseq | 0.0582 | 0.8476 | |
| SRP159526 | VIM | 7431 | RNAseq | 0.1181 | 0.7001 | |
| SRP193095 | VIM | 7431 | RNAseq | 1.4526 | 0.0000 | |
| SRP219564 | VIM | 7431 | RNAseq | 0.3672 | 0.5229 | |
| TCGA | VIM | 7431 | RNAseq | -0.0789 | 0.2729 |
Upregulated datasets: 4; Downregulated datasets: 2.
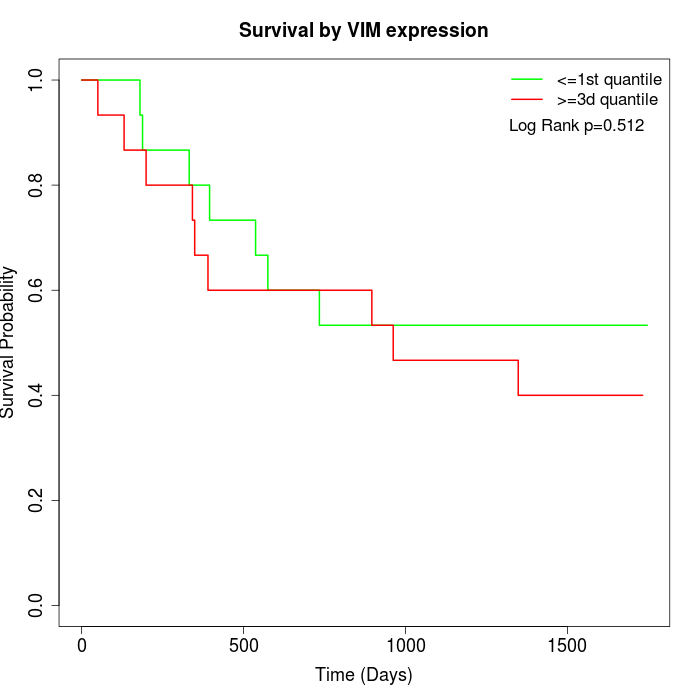
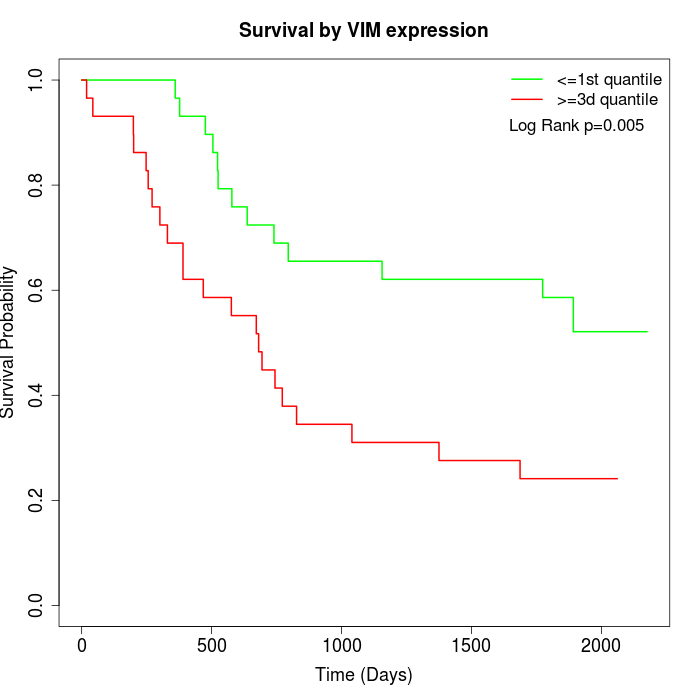
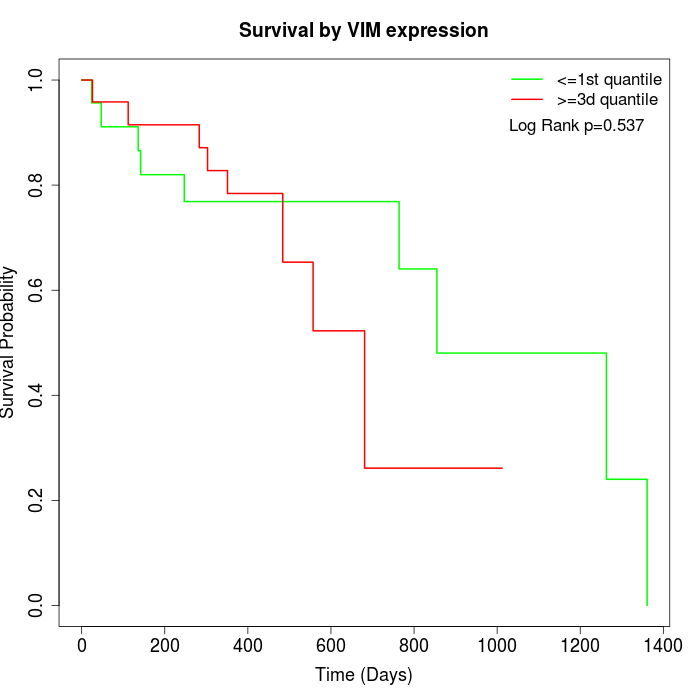
Survival by VIM expression:
Note: Click image to view full size file.
Copy number change of VIM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VIM | 7431 | 3 | 8 | 19 | |
| GSE20123 | VIM | 7431 | 3 | 7 | 20 | |
| GSE43470 | VIM | 7431 | 3 | 4 | 36 | |
| GSE46452 | VIM | 7431 | 1 | 14 | 44 | |
| GSE47630 | VIM | 7431 | 5 | 15 | 20 | |
| GSE54993 | VIM | 7431 | 10 | 0 | 60 | |
| GSE54994 | VIM | 7431 | 4 | 9 | 40 | |
| GSE60625 | VIM | 7431 | 0 | 0 | 11 | |
| GSE74703 | VIM | 7431 | 2 | 3 | 31 | |
| GSE74704 | VIM | 7431 | 0 | 6 | 14 | |
| TCGA | VIM | 7431 | 19 | 26 | 51 |
Total number of gains: 50; Total number of losses: 92; Total Number of normals: 346.
Somatic mutations of VIM:
Generating mutation plots.
Highly correlated genes for VIM:
Showing top 20/854 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VIM | FN1 | 0.768854 | 12 | 0 | 12 |
| VIM | DNM3OS | 0.764312 | 3 | 0 | 3 |
| VIM | FBN1 | 0.760239 | 11 | 0 | 10 |
| VIM | PMP22 | 0.740942 | 12 | 0 | 10 |
| VIM | FSTL1 | 0.732086 | 13 | 0 | 13 |
| VIM | MCAM | 0.723953 | 8 | 0 | 8 |
| VIM | GNG2 | 0.723586 | 4 | 0 | 3 |
| VIM | BICC1 | 0.720019 | 9 | 0 | 9 |
| VIM | TTC28 | 0.719801 | 4 | 0 | 4 |
| VIM | IGFBP7 | 0.712012 | 13 | 0 | 11 |
| VIM | CXCL5 | 0.709307 | 3 | 0 | 3 |
| VIM | COL12A1 | 0.7061 | 9 | 0 | 9 |
| VIM | COL6A2 | 0.703556 | 12 | 0 | 11 |
| VIM | CALD1 | 0.701487 | 12 | 0 | 11 |
| VIM | AEBP1 | 0.70117 | 13 | 0 | 11 |
| VIM | COL6A1 | 0.700955 | 12 | 0 | 11 |
| VIM | LAMA4 | 0.697482 | 12 | 0 | 11 |
| VIM | EMP3 | 0.69605 | 12 | 0 | 12 |
| VIM | NID2 | 0.696043 | 10 | 0 | 10 |
| VIM | C8orf88 | 0.695082 | 3 | 0 | 3 |
For details and further investigation, click here