| Full name: VPS18 core subunit of CORVET and HOPS complexes | Alias Symbol: KIAA1475|PEP3 | ||
| Type: protein-coding gene | Cytoband: 15q15.1 | ||
| Entrez ID: 57617 | HGNC ID: HGNC:15972 | Ensembl Gene: ENSG00000104142 | OMIM ID: 608551 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of VPS18:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VPS18 | 57617 | 223346_at | -0.0418 | 0.9394 | |
| GSE26886 | VPS18 | 57617 | 223346_at | -0.6082 | 0.0053 | |
| GSE45670 | VPS18 | 57617 | 223346_at | 0.2223 | 0.0343 | |
| GSE63941 | VPS18 | 57617 | 223346_at | -1.3791 | 0.0061 | |
| GSE77861 | VPS18 | 57617 | 223346_at | -0.0135 | 0.9441 | |
| GSE97050 | VPS18 | 57617 | A_24_P18802 | -0.0203 | 0.9442 | |
| SRP007169 | VPS18 | 57617 | RNAseq | -0.7524 | 0.0556 | |
| SRP008496 | VPS18 | 57617 | RNAseq | -0.8151 | 0.0014 | |
| SRP064894 | VPS18 | 57617 | RNAseq | 0.2701 | 0.3262 | |
| SRP133303 | VPS18 | 57617 | RNAseq | 0.1931 | 0.1398 | |
| SRP159526 | VPS18 | 57617 | RNAseq | -0.1203 | 0.7128 | |
| SRP193095 | VPS18 | 57617 | RNAseq | 0.2380 | 0.0237 | |
| SRP219564 | VPS18 | 57617 | RNAseq | -0.2079 | 0.6532 | |
| TCGA | VPS18 | 57617 | RNAseq | -0.1091 | 0.0402 |
Upregulated datasets: 0; Downregulated datasets: 1.
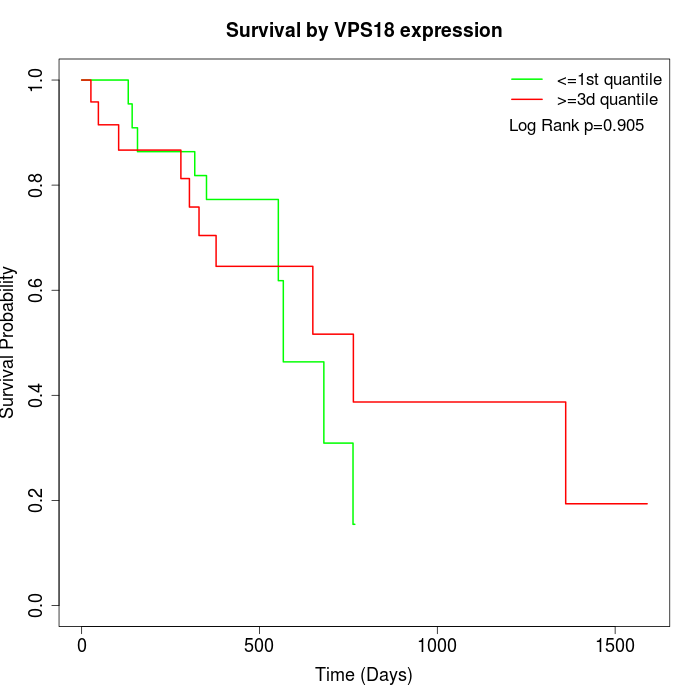
Survival by VPS18 expression:
Note: Click image to view full size file.
Copy number change of VPS18:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VPS18 | 57617 | 3 | 6 | 21 | |
| GSE20123 | VPS18 | 57617 | 3 | 6 | 21 | |
| GSE43470 | VPS18 | 57617 | 4 | 6 | 33 | |
| GSE46452 | VPS18 | 57617 | 3 | 7 | 49 | |
| GSE47630 | VPS18 | 57617 | 8 | 11 | 21 | |
| GSE54993 | VPS18 | 57617 | 4 | 6 | 60 | |
| GSE54994 | VPS18 | 57617 | 6 | 7 | 40 | |
| GSE60625 | VPS18 | 57617 | 4 | 0 | 7 | |
| GSE74703 | VPS18 | 57617 | 4 | 5 | 27 | |
| GSE74704 | VPS18 | 57617 | 2 | 4 | 14 | |
| TCGA | VPS18 | 57617 | 11 | 18 | 67 |
Total number of gains: 52; Total number of losses: 76; Total Number of normals: 360.
Somatic mutations of VPS18:
Generating mutation plots.
Highly correlated genes for VPS18:
Showing top 20/123 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VPS18 | TSPAN3 | 0.755404 | 3 | 0 | 3 |
| VPS18 | PTTG1IP | 0.740884 | 3 | 0 | 3 |
| VPS18 | AIFM1 | 0.729657 | 3 | 0 | 3 |
| VPS18 | ACSS3 | 0.727689 | 3 | 0 | 3 |
| VPS18 | MKRN1 | 0.72246 | 3 | 0 | 3 |
| VPS18 | HIPK1 | 0.720253 | 3 | 0 | 3 |
| VPS18 | CDIPT | 0.712628 | 3 | 0 | 3 |
| VPS18 | TOMM22 | 0.707646 | 3 | 0 | 3 |
| VPS18 | FAM98B | 0.706851 | 3 | 0 | 3 |
| VPS18 | MAP1A | 0.70341 | 3 | 0 | 3 |
| VPS18 | AVEN | 0.699204 | 3 | 0 | 3 |
| VPS18 | NT5E | 0.694136 | 3 | 0 | 3 |
| VPS18 | CSDE1 | 0.681833 | 3 | 0 | 3 |
| VPS18 | SPPL2A | 0.680715 | 3 | 0 | 3 |
| VPS18 | WHAMM | 0.679997 | 3 | 0 | 3 |
| VPS18 | NSMCE1 | 0.677495 | 3 | 0 | 3 |
| VPS18 | MTMR3 | 0.673677 | 3 | 0 | 3 |
| VPS18 | CTTNBP2NL | 0.673367 | 3 | 0 | 3 |
| VPS18 | KIRREL3 | 0.672463 | 3 | 0 | 3 |
| VPS18 | CISD3 | 0.672417 | 3 | 0 | 3 |
For details and further investigation, click here