| Full name: zinc finger FYVE-type containing 16 | Alias Symbol: KIAA0305|PPP1R69 | ||
| Type: protein-coding gene | Cytoband: 5q14.1 | ||
| Entrez ID: 9765 | HGNC ID: HGNC:20756 | Ensembl Gene: ENSG00000039319 | OMIM ID: 608880 |
| Drug and gene relationship at DGIdb | |||
ZFYVE16 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04350 | TGF-beta signaling pathway |
Expression of ZFYVE16:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZFYVE16 | 9765 | 203651_at | -0.0455 | 0.9243 | |
| GSE20347 | ZFYVE16 | 9765 | 203651_at | -0.3904 | 0.0049 | |
| GSE23400 | ZFYVE16 | 9765 | 203651_at | 0.0105 | 0.9039 | |
| GSE26886 | ZFYVE16 | 9765 | 203651_at | -0.4146 | 0.1769 | |
| GSE29001 | ZFYVE16 | 9765 | 203651_at | 0.1206 | 0.6797 | |
| GSE38129 | ZFYVE16 | 9765 | 203651_at | -0.1970 | 0.0828 | |
| GSE45670 | ZFYVE16 | 9765 | 203651_at | -0.0335 | 0.8057 | |
| GSE53622 | ZFYVE16 | 9765 | 19082 | -0.0858 | 0.3393 | |
| GSE53624 | ZFYVE16 | 9765 | 19082 | -0.1540 | 0.0291 | |
| GSE63941 | ZFYVE16 | 9765 | 203651_at | -1.3952 | 0.0004 | |
| GSE77861 | ZFYVE16 | 9765 | 203651_at | -0.2308 | 0.4065 | |
| GSE97050 | ZFYVE16 | 9765 | A_24_P286054 | 0.1656 | 0.4907 | |
| SRP007169 | ZFYVE16 | 9765 | RNAseq | 0.1711 | 0.6299 | |
| SRP008496 | ZFYVE16 | 9765 | RNAseq | 0.1926 | 0.3049 | |
| SRP064894 | ZFYVE16 | 9765 | RNAseq | 0.0913 | 0.6998 | |
| SRP133303 | ZFYVE16 | 9765 | RNAseq | 0.2108 | 0.1921 | |
| SRP159526 | ZFYVE16 | 9765 | RNAseq | -0.2261 | 0.4455 | |
| SRP193095 | ZFYVE16 | 9765 | RNAseq | -0.3272 | 0.0002 | |
| SRP219564 | ZFYVE16 | 9765 | RNAseq | -0.3627 | 0.1739 | |
| TCGA | ZFYVE16 | 9765 | RNAseq | -0.1000 | 0.0439 |
Upregulated datasets: 0; Downregulated datasets: 1.
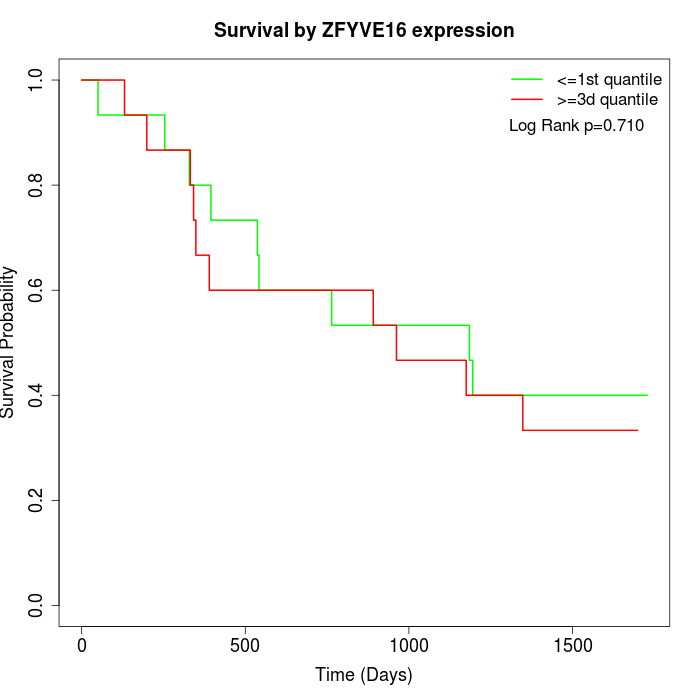
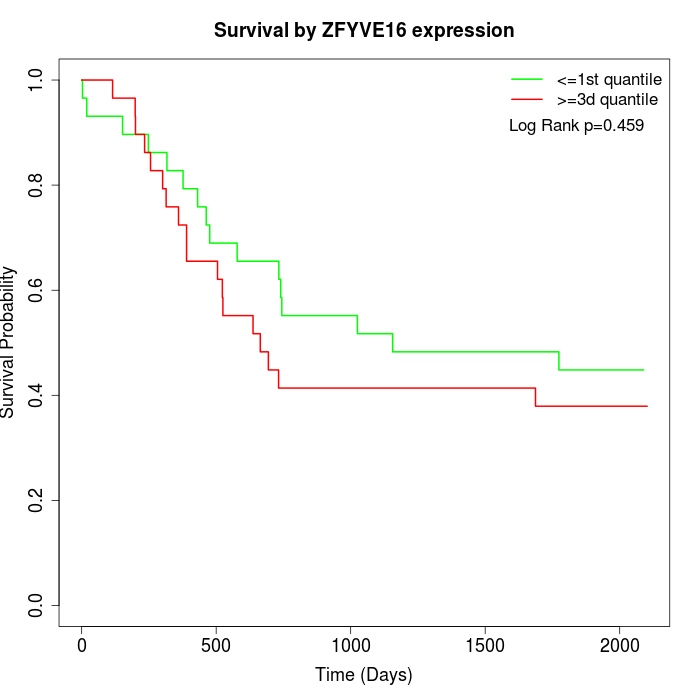
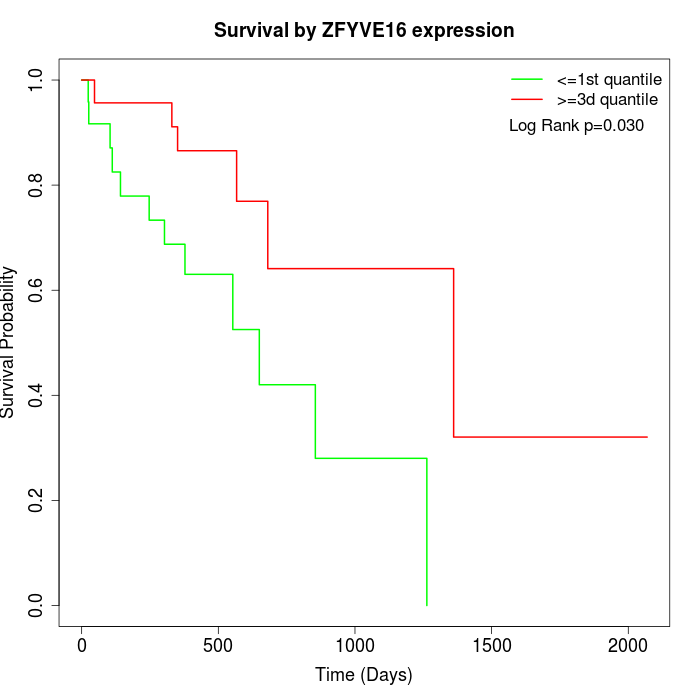
Survival by ZFYVE16 expression:
Note: Click image to view full size file.
Copy number change of ZFYVE16:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZFYVE16 | 9765 | 1 | 13 | 16 | |
| GSE20123 | ZFYVE16 | 9765 | 1 | 13 | 16 | |
| GSE43470 | ZFYVE16 | 9765 | 1 | 11 | 31 | |
| GSE46452 | ZFYVE16 | 9765 | 0 | 28 | 31 | |
| GSE47630 | ZFYVE16 | 9765 | 1 | 19 | 20 | |
| GSE54993 | ZFYVE16 | 9765 | 8 | 1 | 61 | |
| GSE54994 | ZFYVE16 | 9765 | 2 | 18 | 33 | |
| GSE60625 | ZFYVE16 | 9765 | 0 | 0 | 11 | |
| GSE74703 | ZFYVE16 | 9765 | 1 | 7 | 28 | |
| GSE74704 | ZFYVE16 | 9765 | 1 | 6 | 13 | |
| TCGA | ZFYVE16 | 9765 | 5 | 49 | 42 |
Total number of gains: 21; Total number of losses: 165; Total Number of normals: 302.
Somatic mutations of ZFYVE16:
Generating mutation plots.
Highly correlated genes for ZFYVE16:
Showing top 20/349 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZFYVE16 | ARL10 | 0.747081 | 3 | 0 | 3 |
| ZFYVE16 | TRAPPC6B | 0.735304 | 3 | 0 | 3 |
| ZFYVE16 | ATG2B | 0.73277 | 5 | 0 | 5 |
| ZFYVE16 | COG3 | 0.726113 | 3 | 0 | 3 |
| ZFYVE16 | BDKRB1 | 0.718765 | 3 | 0 | 3 |
| ZFYVE16 | PLIN2 | 0.712648 | 3 | 0 | 3 |
| ZFYVE16 | YAF2 | 0.709299 | 3 | 0 | 3 |
| ZFYVE16 | SNX18 | 0.70028 | 4 | 0 | 4 |
| ZFYVE16 | BDP1 | 0.699333 | 8 | 0 | 7 |
| ZFYVE16 | ZNF600 | 0.696259 | 3 | 0 | 3 |
| ZFYVE16 | VSTM4 | 0.696209 | 3 | 0 | 3 |
| ZFYVE16 | FAM174A | 0.692789 | 4 | 0 | 4 |
| ZFYVE16 | FAM193B | 0.692416 | 3 | 0 | 3 |
| ZFYVE16 | DHRS7 | 0.688706 | 4 | 0 | 3 |
| ZFYVE16 | GFM2 | 0.684069 | 5 | 0 | 4 |
| ZFYVE16 | EAF1 | 0.682003 | 5 | 0 | 5 |
| ZFYVE16 | HOOK3 | 0.681543 | 4 | 0 | 3 |
| ZFYVE16 | ALDH7A1 | 0.678583 | 4 | 0 | 3 |
| ZFYVE16 | SGPP1 | 0.677553 | 4 | 0 | 3 |
| ZFYVE16 | TMEM161B | 0.675269 | 6 | 0 | 5 |
For details and further investigation, click here