| Full name: acyl-CoA thioesterase 9 | Alias Symbol: CGI-16|MT-ACT48|ACATE2 | ||
| Type: protein-coding gene | Cytoband: Xp22.11 | ||
| Entrez ID: 23597 | HGNC ID: HGNC:17152 | Ensembl Gene: ENSG00000123130 | OMIM ID: 300862 |
| Drug and gene relationship at DGIdb | |||
Expression of ACOT9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACOT9 | 23597 | 221641_s_at | 0.4037 | 0.4446 | |
| GSE20347 | ACOT9 | 23597 | 221641_s_at | 1.1648 | 0.0000 | |
| GSE23400 | ACOT9 | 23597 | 221641_s_at | 0.7128 | 0.0000 | |
| GSE26886 | ACOT9 | 23597 | 221641_s_at | 1.4664 | 0.0000 | |
| GSE29001 | ACOT9 | 23597 | 221641_s_at | 1.4333 | 0.0004 | |
| GSE38129 | ACOT9 | 23597 | 221641_s_at | 0.7695 | 0.0024 | |
| GSE45670 | ACOT9 | 23597 | 221641_s_at | 0.3355 | 0.2820 | |
| GSE53622 | ACOT9 | 23597 | 156342 | 0.4591 | 0.0000 | |
| GSE53624 | ACOT9 | 23597 | 156342 | 0.8278 | 0.0000 | |
| GSE63941 | ACOT9 | 23597 | 221641_s_at | -0.6827 | 0.2619 | |
| GSE77861 | ACOT9 | 23597 | 221641_s_at | 0.8824 | 0.0055 | |
| GSE97050 | ACOT9 | 23597 | A_24_P42501 | 0.1936 | 0.3480 | |
| SRP007169 | ACOT9 | 23597 | RNAseq | 1.8779 | 0.0001 | |
| SRP008496 | ACOT9 | 23597 | RNAseq | 1.7359 | 0.0000 | |
| SRP064894 | ACOT9 | 23597 | RNAseq | 0.8904 | 0.0000 | |
| SRP133303 | ACOT9 | 23597 | RNAseq | 1.2395 | 0.0000 | |
| SRP159526 | ACOT9 | 23597 | RNAseq | 0.4775 | 0.0189 | |
| SRP193095 | ACOT9 | 23597 | RNAseq | 1.1974 | 0.0000 | |
| SRP219564 | ACOT9 | 23597 | RNAseq | 0.4900 | 0.2958 | |
| TCGA | ACOT9 | 23597 | RNAseq | 0.0190 | 0.7980 |
Upregulated datasets: 7; Downregulated datasets: 0.
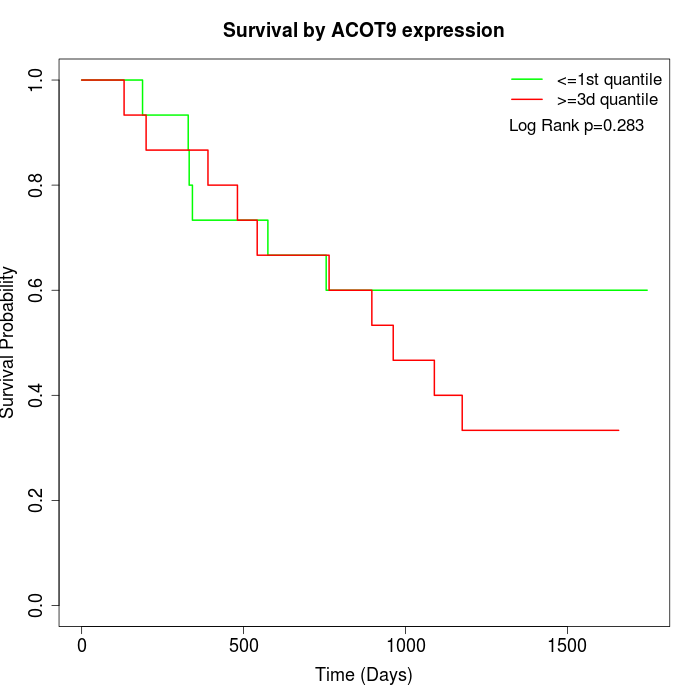
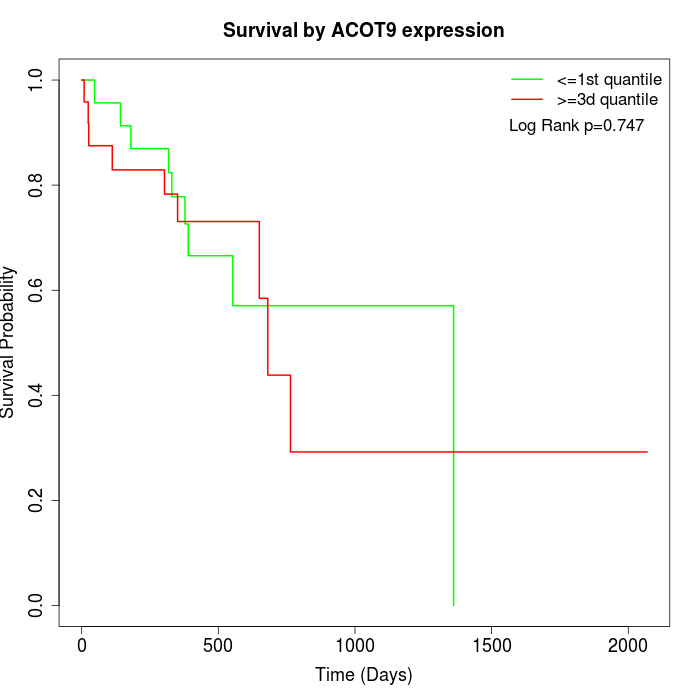
Survival by ACOT9 expression:
Note: Click image to view full size file.
Copy number change of ACOT9:
No record found for this gene.
Somatic mutations of ACOT9:
Generating mutation plots.
Highly correlated genes for ACOT9:
Showing top 20/1198 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACOT9 | INHBA | 0.747983 | 11 | 0 | 10 |
| ACOT9 | GOLM1 | 0.747321 | 6 | 0 | 6 |
| ACOT9 | SERPINH1 | 0.735308 | 11 | 0 | 11 |
| ACOT9 | LGALS1 | 0.728741 | 11 | 0 | 10 |
| ACOT9 | CLIC4 | 0.726942 | 10 | 0 | 10 |
| ACOT9 | COL4A1 | 0.724019 | 11 | 0 | 11 |
| ACOT9 | FNDC3B | 0.723695 | 11 | 0 | 11 |
| ACOT9 | TGFBI | 0.723226 | 12 | 0 | 11 |
| ACOT9 | LOXL2 | 0.720267 | 10 | 0 | 10 |
| ACOT9 | POP7 | 0.705133 | 5 | 0 | 5 |
| ACOT9 | PMEPA1 | 0.70339 | 10 | 0 | 9 |
| ACOT9 | CALU | 0.700844 | 10 | 0 | 10 |
| ACOT9 | ENAH | 0.697881 | 12 | 0 | 11 |
| ACOT9 | MET | 0.694115 | 8 | 0 | 8 |
| ACOT9 | SLC39A14 | 0.693937 | 11 | 0 | 10 |
| ACOT9 | MMD | 0.693401 | 12 | 0 | 11 |
| ACOT9 | COL4A2 | 0.692479 | 11 | 0 | 11 |
| ACOT9 | SEMA3C | 0.68979 | 10 | 0 | 8 |
| ACOT9 | LOXL1-AS1 | 0.689477 | 5 | 0 | 5 |
| ACOT9 | PLAUR | 0.687336 | 11 | 0 | 11 |
For details and further investigation, click here