| Full name: anaphase promoting complex subunit 2 | Alias Symbol: APC2|KIAA1406 | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 29882 | HGNC ID: HGNC:19989 | Ensembl Gene: ENSG00000176248 | OMIM ID: 606946 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ANAPC2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04914 | Progesterone-mediated oocyte maturation | |
| hsa05166 | HTLV-I infection |
Expression of ANAPC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ANAPC2 | 29882 | 218555_at | 0.2841 | 0.5843 | |
| GSE20347 | ANAPC2 | 29882 | 218555_at | -0.0640 | 0.5879 | |
| GSE23400 | ANAPC2 | 29882 | 218555_at | 0.1303 | 0.0199 | |
| GSE26886 | ANAPC2 | 29882 | 218555_at | 0.7685 | 0.0000 | |
| GSE29001 | ANAPC2 | 29882 | 218555_at | 0.0864 | 0.6032 | |
| GSE38129 | ANAPC2 | 29882 | 218555_at | 0.0083 | 0.9361 | |
| GSE45670 | ANAPC2 | 29882 | 218555_at | 0.1457 | 0.1113 | |
| GSE53622 | ANAPC2 | 29882 | 80090 | -0.0640 | 0.3742 | |
| GSE53624 | ANAPC2 | 29882 | 80090 | 0.1335 | 0.0616 | |
| GSE63941 | ANAPC2 | 29882 | 218555_at | 1.7379 | 0.0000 | |
| GSE77861 | ANAPC2 | 29882 | 218555_at | 0.1294 | 0.3604 | |
| GSE97050 | ANAPC2 | 29882 | A_23_P253068 | 0.1359 | 0.6575 | |
| SRP007169 | ANAPC2 | 29882 | RNAseq | -0.3187 | 0.4319 | |
| SRP008496 | ANAPC2 | 29882 | RNAseq | -0.4830 | 0.1008 | |
| SRP064894 | ANAPC2 | 29882 | RNAseq | 0.4662 | 0.0173 | |
| SRP133303 | ANAPC2 | 29882 | RNAseq | -0.2003 | 0.2271 | |
| SRP159526 | ANAPC2 | 29882 | RNAseq | 0.1240 | 0.5656 | |
| SRP193095 | ANAPC2 | 29882 | RNAseq | 0.1991 | 0.2491 | |
| SRP219564 | ANAPC2 | 29882 | RNAseq | 0.3154 | 0.4908 | |
| TCGA | ANAPC2 | 29882 | RNAseq | -0.0378 | 0.4494 |
Upregulated datasets: 1; Downregulated datasets: 0.
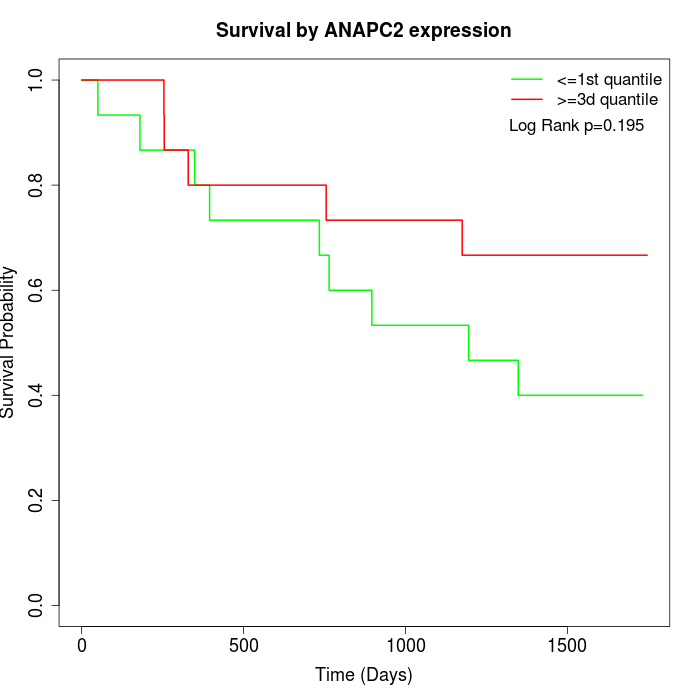
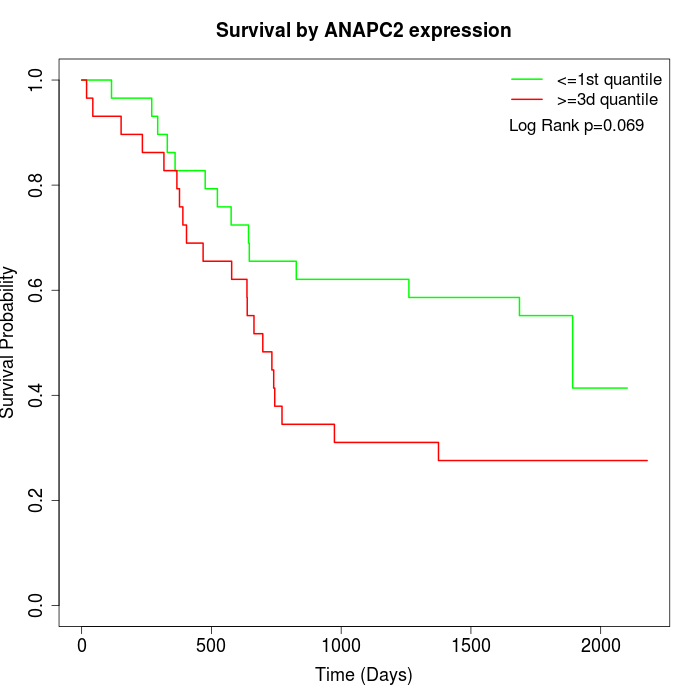
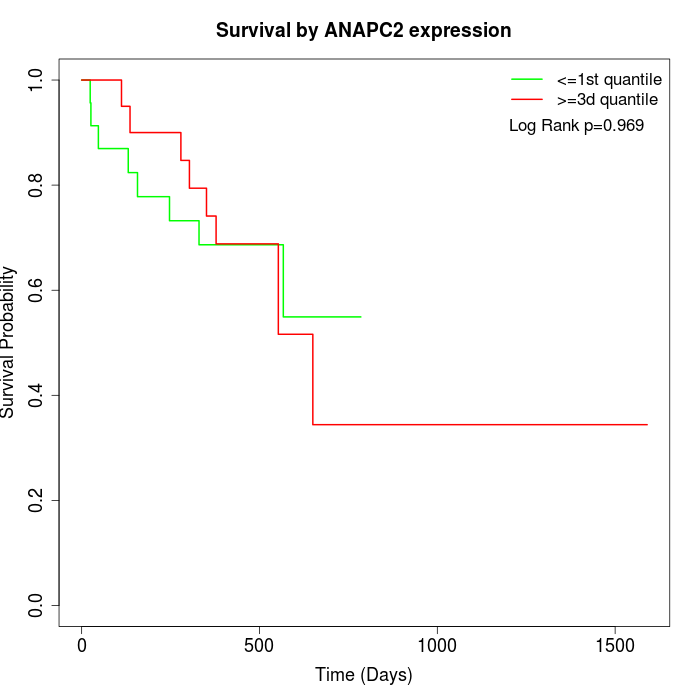
Survival by ANAPC2 expression:
Note: Click image to view full size file.
Copy number change of ANAPC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ANAPC2 | 29882 | 5 | 7 | 18 | |
| GSE20123 | ANAPC2 | 29882 | 5 | 7 | 18 | |
| GSE43470 | ANAPC2 | 29882 | 3 | 7 | 33 | |
| GSE46452 | ANAPC2 | 29882 | 6 | 13 | 40 | |
| GSE47630 | ANAPC2 | 29882 | 6 | 15 | 19 | |
| GSE54993 | ANAPC2 | 29882 | 3 | 3 | 64 | |
| GSE54994 | ANAPC2 | 29882 | 12 | 8 | 33 | |
| GSE60625 | ANAPC2 | 29882 | 0 | 0 | 11 | |
| GSE74703 | ANAPC2 | 29882 | 3 | 5 | 28 | |
| GSE74704 | ANAPC2 | 29882 | 3 | 5 | 12 | |
| TCGA | ANAPC2 | 29882 | 29 | 23 | 44 |
Total number of gains: 75; Total number of losses: 93; Total Number of normals: 320.
Somatic mutations of ANAPC2:
Generating mutation plots.
Highly correlated genes for ANAPC2:
Showing top 20/315 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ANAPC2 | MTFP1 | 0.688632 | 3 | 0 | 3 |
| ANAPC2 | BSG | 0.684365 | 3 | 0 | 3 |
| ANAPC2 | ANGPTL3 | 0.662538 | 3 | 0 | 3 |
| ANAPC2 | SLC27A1 | 0.661557 | 4 | 0 | 4 |
| ANAPC2 | SIGLEC9 | 0.653818 | 3 | 0 | 3 |
| ANAPC2 | UBOX5 | 0.647075 | 3 | 0 | 3 |
| ANAPC2 | NOP2 | 0.646178 | 3 | 0 | 3 |
| ANAPC2 | SIGLEC7 | 0.643765 | 3 | 0 | 3 |
| ANAPC2 | NELFB | 0.643425 | 10 | 0 | 9 |
| ANAPC2 | LENEP | 0.64035 | 3 | 0 | 3 |
| ANAPC2 | TSPAN32 | 0.639711 | 4 | 0 | 4 |
| ANAPC2 | RTEL1 | 0.638628 | 3 | 0 | 3 |
| ANAPC2 | TJAP1 | 0.637632 | 3 | 0 | 3 |
| ANAPC2 | RABL6 | 0.637563 | 5 | 0 | 4 |
| ANAPC2 | NXPH4 | 0.636606 | 5 | 0 | 5 |
| ANAPC2 | LILRA3 | 0.634486 | 4 | 0 | 4 |
| ANAPC2 | TRAPPC1 | 0.634185 | 4 | 0 | 3 |
| ANAPC2 | IRF3 | 0.632827 | 5 | 0 | 3 |
| ANAPC2 | HS6ST2 | 0.629032 | 3 | 0 | 3 |
| ANAPC2 | SNX10 | 0.628895 | 3 | 0 | 3 |
For details and further investigation, click here