| Full name: Rho GTPase activating protein 29 | Alias Symbol: PARG1 | ||
| Type: protein-coding gene | Cytoband: 1p22.1 | ||
| Entrez ID: 9411 | HGNC ID: HGNC:30207 | Ensembl Gene: ENSG00000137962 | OMIM ID: 610496 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ARHGAP29:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGAP29 | 9411 | 203910_at | -0.4003 | 0.4050 | |
| GSE20347 | ARHGAP29 | 9411 | 203910_at | -0.0063 | 0.9862 | |
| GSE23400 | ARHGAP29 | 9411 | 203910_at | 0.0843 | 0.5693 | |
| GSE26886 | ARHGAP29 | 9411 | 203910_at | 0.1937 | 0.7810 | |
| GSE29001 | ARHGAP29 | 9411 | 203910_at | 0.2205 | 0.6184 | |
| GSE38129 | ARHGAP29 | 9411 | 203910_at | -0.2141 | 0.5332 | |
| GSE45670 | ARHGAP29 | 9411 | 203910_at | -0.9675 | 0.0376 | |
| GSE53622 | ARHGAP29 | 9411 | 79824 | -0.2151 | 0.1582 | |
| GSE53624 | ARHGAP29 | 9411 | 79824 | -0.0631 | 0.5691 | |
| GSE63941 | ARHGAP29 | 9411 | 203910_at | 0.1918 | 0.8991 | |
| GSE77861 | ARHGAP29 | 9411 | 203910_at | 0.9817 | 0.0822 | |
| GSE97050 | ARHGAP29 | 9411 | A_24_P333571 | 0.4119 | 0.3428 | |
| SRP007169 | ARHGAP29 | 9411 | RNAseq | 0.2284 | 0.5968 | |
| SRP008496 | ARHGAP29 | 9411 | RNAseq | 0.7628 | 0.0171 | |
| SRP064894 | ARHGAP29 | 9411 | RNAseq | 0.3426 | 0.2735 | |
| SRP133303 | ARHGAP29 | 9411 | RNAseq | 0.1444 | 0.6274 | |
| SRP159526 | ARHGAP29 | 9411 | RNAseq | -0.1068 | 0.7611 | |
| SRP193095 | ARHGAP29 | 9411 | RNAseq | 1.1641 | 0.0004 | |
| SRP219564 | ARHGAP29 | 9411 | RNAseq | 0.1253 | 0.8118 | |
| TCGA | ARHGAP29 | 9411 | RNAseq | -0.4145 | 0.0019 |
Upregulated datasets: 1; Downregulated datasets: 0.
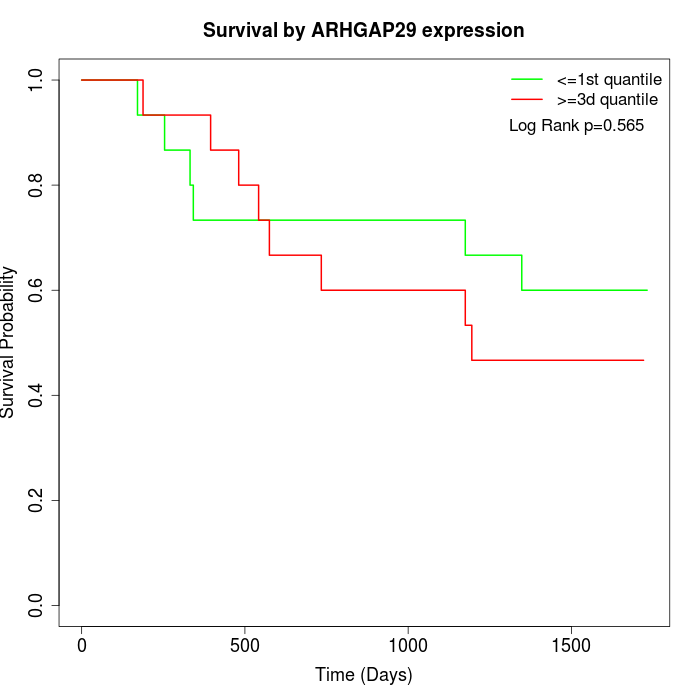
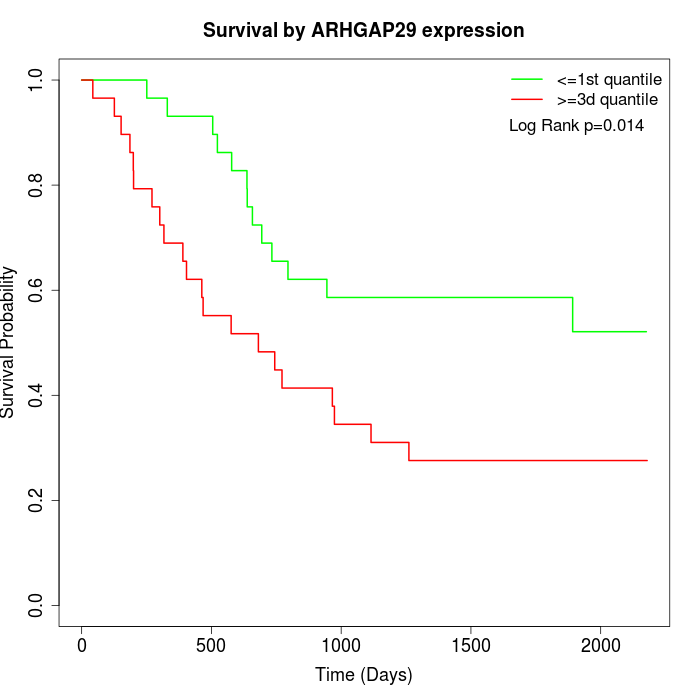
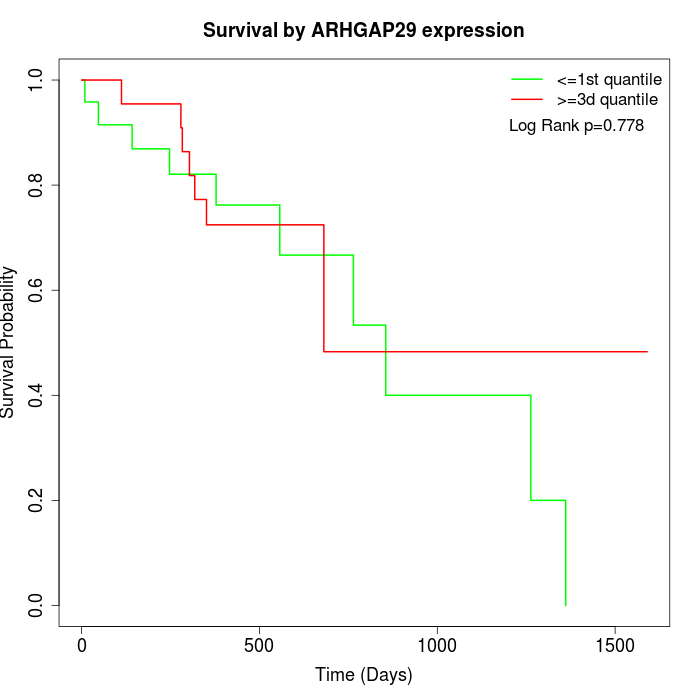
Survival by ARHGAP29 expression:
Note: Click image to view full size file.
Copy number change of ARHGAP29:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGAP29 | 9411 | 0 | 8 | 22 | |
| GSE20123 | ARHGAP29 | 9411 | 0 | 8 | 22 | |
| GSE43470 | ARHGAP29 | 9411 | 2 | 6 | 35 | |
| GSE46452 | ARHGAP29 | 9411 | 1 | 1 | 57 | |
| GSE47630 | ARHGAP29 | 9411 | 8 | 5 | 27 | |
| GSE54993 | ARHGAP29 | 9411 | 0 | 1 | 69 | |
| GSE54994 | ARHGAP29 | 9411 | 6 | 3 | 44 | |
| GSE60625 | ARHGAP29 | 9411 | 0 | 0 | 11 | |
| GSE74703 | ARHGAP29 | 9411 | 1 | 5 | 30 | |
| GSE74704 | ARHGAP29 | 9411 | 0 | 5 | 15 | |
| TCGA | ARHGAP29 | 9411 | 6 | 24 | 66 |
Total number of gains: 24; Total number of losses: 66; Total Number of normals: 398.
Somatic mutations of ARHGAP29:
Generating mutation plots.
Highly correlated genes for ARHGAP29:
Showing top 20/354 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGAP29 | CNRIP1 | 0.74446 | 3 | 0 | 3 |
| ARHGAP29 | BVES | 0.70444 | 3 | 0 | 3 |
| ARHGAP29 | LIX1L | 0.70199 | 3 | 0 | 3 |
| ARHGAP29 | BICC1 | 0.69854 | 4 | 0 | 4 |
| ARHGAP29 | ITIH5 | 0.695984 | 3 | 0 | 3 |
| ARHGAP29 | SYNE1 | 0.691522 | 4 | 0 | 4 |
| ARHGAP29 | DCHS1 | 0.689217 | 5 | 0 | 4 |
| ARHGAP29 | ZNF423 | 0.686544 | 5 | 0 | 5 |
| ARHGAP29 | OLFML1 | 0.683183 | 5 | 0 | 5 |
| ARHGAP29 | IGDCC4 | 0.682319 | 3 | 0 | 3 |
| ARHGAP29 | KCTD12 | 0.6809 | 5 | 0 | 5 |
| ARHGAP29 | EPB41L2 | 0.677117 | 4 | 0 | 4 |
| ARHGAP29 | JAM2 | 0.67606 | 4 | 0 | 4 |
| ARHGAP29 | FILIP1L | 0.67391 | 7 | 0 | 6 |
| ARHGAP29 | SLIT3 | 0.671119 | 3 | 0 | 3 |
| ARHGAP29 | COL6A1 | 0.669994 | 5 | 0 | 5 |
| ARHGAP29 | DDR2 | 0.668888 | 5 | 0 | 5 |
| ARHGAP29 | SFRP2 | 0.668301 | 3 | 0 | 3 |
| ARHGAP29 | ENPP2 | 0.667099 | 5 | 0 | 5 |
| ARHGAP29 | SDC2 | 0.662997 | 6 | 0 | 5 |
For details and further investigation, click here