| Full name: mitochondrial translational initiation factor 2 | Alias Symbol: IF-2mt | ||
| Type: protein-coding gene | Cytoband: 2p16.1 | ||
| Entrez ID: 4528 | HGNC ID: HGNC:7441 | Ensembl Gene: ENSG00000085760 | OMIM ID: 603766 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MTIF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MTIF2 | 4528 | 203095_at | -0.0108 | 0.9864 | |
| GSE20347 | MTIF2 | 4528 | 203095_at | 0.3376 | 0.0083 | |
| GSE23400 | MTIF2 | 4528 | 203095_at | 0.5223 | 0.0000 | |
| GSE26886 | MTIF2 | 4528 | 203095_at | -0.0989 | 0.6178 | |
| GSE29001 | MTIF2 | 4528 | 203095_at | 0.4987 | 0.0568 | |
| GSE38129 | MTIF2 | 4528 | 203095_at | 0.4843 | 0.0000 | |
| GSE45670 | MTIF2 | 4528 | 203095_at | 0.4393 | 0.0016 | |
| GSE53622 | MTIF2 | 4528 | 23851 | 0.5283 | 0.0000 | |
| GSE53624 | MTIF2 | 4528 | 23851 | 0.5491 | 0.0000 | |
| GSE63941 | MTIF2 | 4528 | 203095_at | -0.0905 | 0.8300 | |
| GSE77861 | MTIF2 | 4528 | 203095_at | 0.3450 | 0.2527 | |
| GSE97050 | MTIF2 | 4528 | A_23_P40072 | 0.3701 | 0.4342 | |
| SRP007169 | MTIF2 | 4528 | RNAseq | 0.7491 | 0.0420 | |
| SRP008496 | MTIF2 | 4528 | RNAseq | 0.5974 | 0.0322 | |
| SRP064894 | MTIF2 | 4528 | RNAseq | 0.3074 | 0.0310 | |
| SRP133303 | MTIF2 | 4528 | RNAseq | 0.3146 | 0.0948 | |
| SRP159526 | MTIF2 | 4528 | RNAseq | 0.5051 | 0.0088 | |
| SRP193095 | MTIF2 | 4528 | RNAseq | -0.0151 | 0.8881 | |
| SRP219564 | MTIF2 | 4528 | RNAseq | 0.4970 | 0.1855 | |
| TCGA | MTIF2 | 4528 | RNAseq | 0.0582 | 0.2582 |
Upregulated datasets: 0; Downregulated datasets: 0.
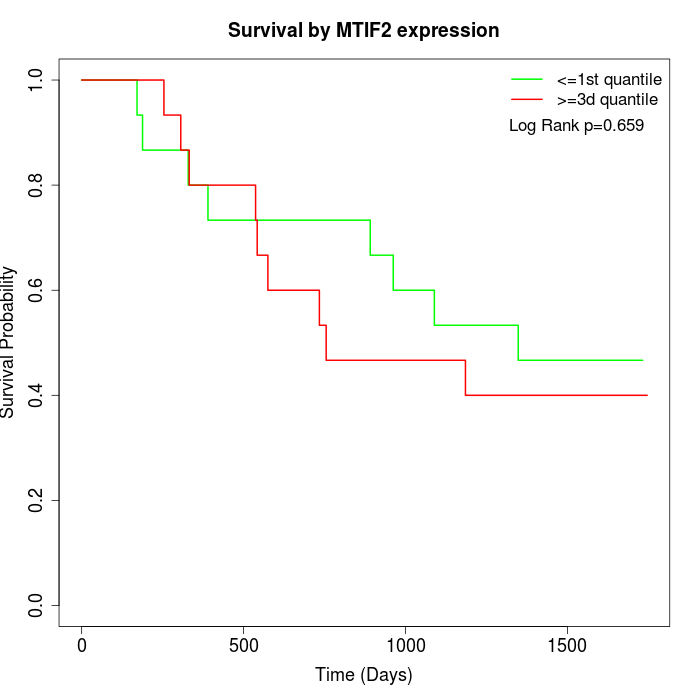
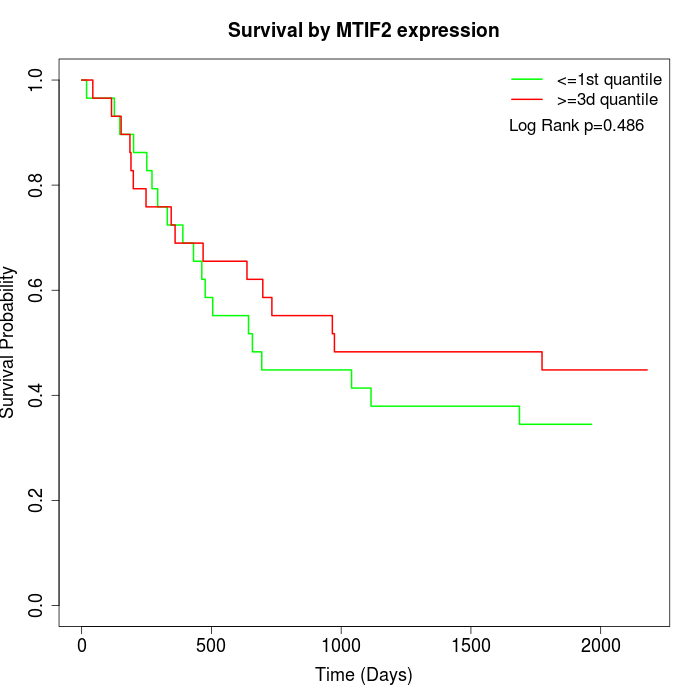
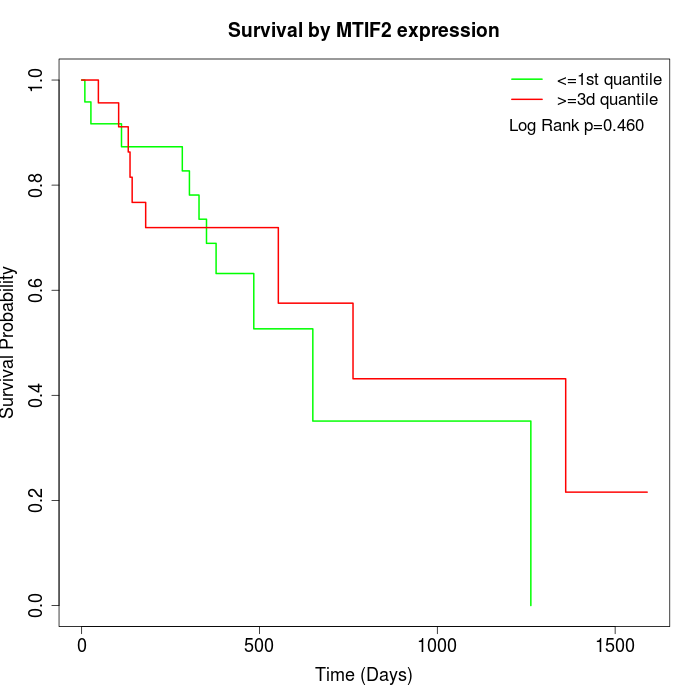
Survival by MTIF2 expression:
Note: Click image to view full size file.
Copy number change of MTIF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MTIF2 | 4528 | 10 | 0 | 20 | |
| GSE20123 | MTIF2 | 4528 | 9 | 0 | 21 | |
| GSE43470 | MTIF2 | 4528 | 5 | 0 | 38 | |
| GSE46452 | MTIF2 | 4528 | 2 | 3 | 54 | |
| GSE47630 | MTIF2 | 4528 | 8 | 0 | 32 | |
| GSE54993 | MTIF2 | 4528 | 0 | 5 | 65 | |
| GSE54994 | MTIF2 | 4528 | 12 | 0 | 41 | |
| GSE60625 | MTIF2 | 4528 | 0 | 3 | 8 | |
| GSE74703 | MTIF2 | 4528 | 5 | 0 | 31 | |
| GSE74704 | MTIF2 | 4528 | 9 | 0 | 11 | |
| TCGA | MTIF2 | 4528 | 34 | 3 | 59 |
Total number of gains: 94; Total number of losses: 14; Total Number of normals: 380.
Somatic mutations of MTIF2:
Generating mutation plots.
Highly correlated genes for MTIF2:
Showing top 20/1160 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MTIF2 | IPMK | 0.789729 | 3 | 0 | 3 |
| MTIF2 | MAP4K5 | 0.776622 | 3 | 0 | 3 |
| MTIF2 | C11orf80 | 0.733404 | 3 | 0 | 3 |
| MTIF2 | PVT1 | 0.731654 | 3 | 0 | 3 |
| MTIF2 | FAF2 | 0.7269 | 3 | 0 | 3 |
| MTIF2 | ISCA2 | 0.724415 | 3 | 0 | 3 |
| MTIF2 | LSM4 | 0.717522 | 5 | 0 | 5 |
| MTIF2 | LAP3 | 0.717457 | 3 | 0 | 3 |
| MTIF2 | PPWD1 | 0.717031 | 3 | 0 | 3 |
| MTIF2 | AGFG1 | 0.716928 | 3 | 0 | 3 |
| MTIF2 | PPP6R3 | 0.71183 | 3 | 0 | 3 |
| MTIF2 | SLC19A1 | 0.706987 | 3 | 0 | 3 |
| MTIF2 | ACP1 | 0.706472 | 8 | 0 | 7 |
| MTIF2 | ATG12 | 0.697458 | 3 | 0 | 3 |
| MTIF2 | HPS5 | 0.693861 | 3 | 0 | 3 |
| MTIF2 | CAD | 0.690881 | 8 | 0 | 8 |
| MTIF2 | TPRA1 | 0.687126 | 3 | 0 | 3 |
| MTIF2 | RANBP6 | 0.686835 | 3 | 0 | 3 |
| MTIF2 | TPX2 | 0.683251 | 8 | 0 | 8 |
| MTIF2 | NEK2 | 0.680828 | 8 | 0 | 8 |
For details and further investigation, click here