| Full name: BCL2 antagonist/killer 1 | Alias Symbol: BCL2L7|BAK | ||
| Type: protein-coding gene | Cytoband: 6p21.31 | ||
| Entrez ID: 578 | HGNC ID: HGNC:949 | Ensembl Gene: ENSG00000030110 | OMIM ID: 600516 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
BAK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04210 | Apoptosis |
Expression of BAK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BAK1 | 578 | 203728_at | 0.9009 | 0.0503 | |
| GSE20347 | BAK1 | 578 | 203728_at | 0.1441 | 0.2791 | |
| GSE23400 | BAK1 | 578 | 203728_at | 0.3084 | 0.0000 | |
| GSE26886 | BAK1 | 578 | 203728_at | -0.4187 | 0.0315 | |
| GSE29001 | BAK1 | 578 | 203728_at | 0.3852 | 0.1640 | |
| GSE38129 | BAK1 | 578 | 203728_at | 0.3992 | 0.0010 | |
| GSE45670 | BAK1 | 578 | 203728_at | 0.7362 | 0.0002 | |
| GSE53622 | BAK1 | 578 | 78148 | 0.5459 | 0.0000 | |
| GSE53624 | BAK1 | 578 | 78148 | 0.7197 | 0.0000 | |
| GSE63941 | BAK1 | 578 | 203728_at | 0.4612 | 0.1894 | |
| GSE77861 | BAK1 | 578 | 203728_at | 0.0988 | 0.6507 | |
| GSE97050 | BAK1 | 578 | A_23_P145357 | 0.2172 | 0.5193 | |
| SRP007169 | BAK1 | 578 | RNAseq | 0.2164 | 0.6160 | |
| SRP008496 | BAK1 | 578 | RNAseq | 0.2000 | 0.4369 | |
| SRP064894 | BAK1 | 578 | RNAseq | 0.5386 | 0.0155 | |
| SRP133303 | BAK1 | 578 | RNAseq | 0.8070 | 0.0053 | |
| SRP159526 | BAK1 | 578 | RNAseq | 0.2636 | 0.4962 | |
| SRP193095 | BAK1 | 578 | RNAseq | 0.1751 | 0.3412 | |
| SRP219564 | BAK1 | 578 | RNAseq | 0.3479 | 0.5459 | |
| TCGA | BAK1 | 578 | RNAseq | 0.2608 | 0.0010 |
Upregulated datasets: 0; Downregulated datasets: 0.
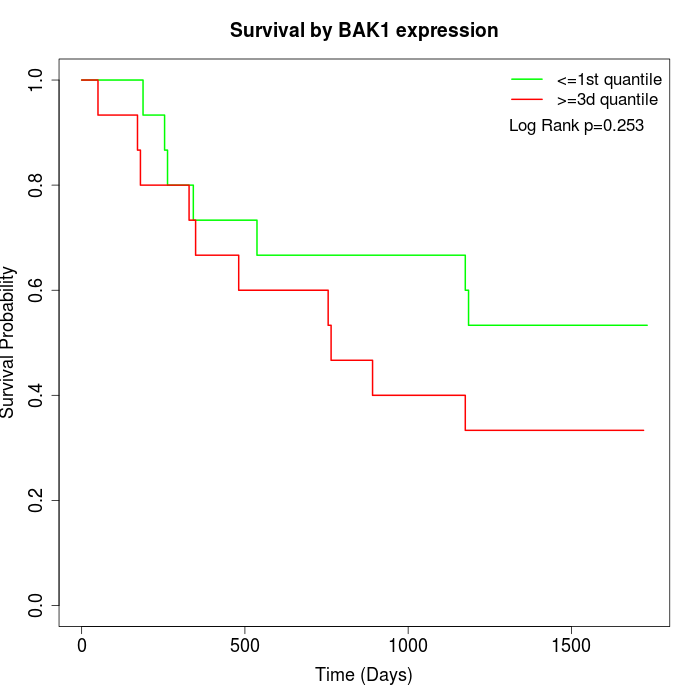
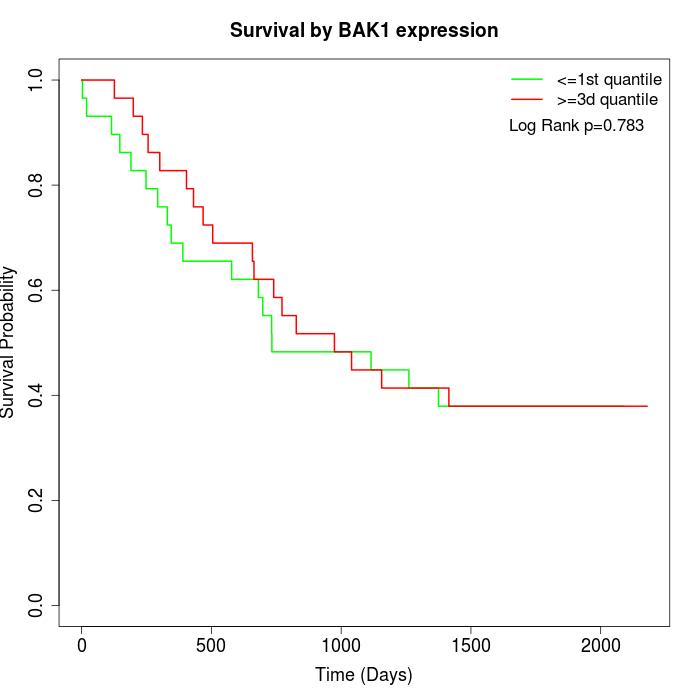
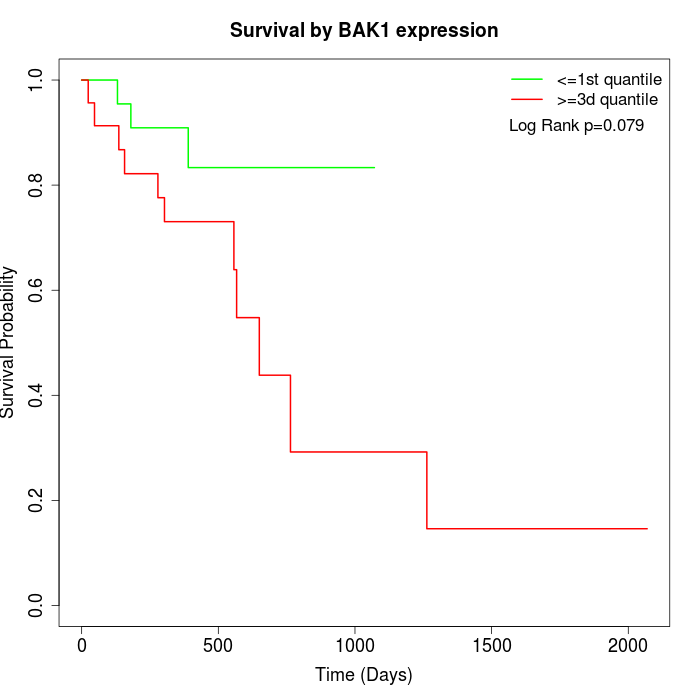
Survival by BAK1 expression:
Note: Click image to view full size file.
Copy number change of BAK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BAK1 | 578 | 5 | 0 | 25 | |
| GSE20123 | BAK1 | 578 | 5 | 0 | 25 | |
| GSE43470 | BAK1 | 578 | 4 | 1 | 38 | |
| GSE46452 | BAK1 | 578 | 2 | 9 | 48 | |
| GSE47630 | BAK1 | 578 | 8 | 4 | 28 | |
| GSE54993 | BAK1 | 578 | 3 | 1 | 66 | |
| GSE54994 | BAK1 | 578 | 11 | 4 | 38 | |
| GSE60625 | BAK1 | 578 | 1 | 0 | 10 | |
| GSE74703 | BAK1 | 578 | 4 | 0 | 32 | |
| GSE74704 | BAK1 | 578 | 2 | 0 | 18 | |
| TCGA | BAK1 | 578 | 15 | 16 | 65 |
Total number of gains: 60; Total number of losses: 35; Total Number of normals: 393.
Somatic mutations of BAK1:
Generating mutation plots.
Highly correlated genes for BAK1:
Showing top 20/638 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BAK1 | MOB3C | 0.851633 | 3 | 0 | 3 |
| BAK1 | GIT1 | 0.778482 | 3 | 0 | 3 |
| BAK1 | RAB12 | 0.749754 | 3 | 0 | 3 |
| BAK1 | FBXO33 | 0.746459 | 3 | 0 | 3 |
| BAK1 | UBR7 | 0.74293 | 4 | 0 | 3 |
| BAK1 | ERP27 | 0.741242 | 3 | 0 | 3 |
| BAK1 | KIAA0586 | 0.740711 | 3 | 0 | 3 |
| BAK1 | TBC1D16 | 0.738614 | 3 | 0 | 3 |
| BAK1 | PRDM15 | 0.736695 | 3 | 0 | 3 |
| BAK1 | ROMO1 | 0.736075 | 3 | 0 | 3 |
| BAK1 | PPP1R9B | 0.731628 | 3 | 0 | 3 |
| BAK1 | HTRA2 | 0.724453 | 3 | 0 | 3 |
| BAK1 | KDM1B | 0.718261 | 3 | 0 | 3 |
| BAK1 | JTB | 0.71303 | 3 | 0 | 3 |
| BAK1 | AGPAT3 | 0.710082 | 3 | 0 | 3 |
| BAK1 | CHST3 | 0.708228 | 3 | 0 | 3 |
| BAK1 | GUK1 | 0.706063 | 3 | 0 | 3 |
| BAK1 | KHNYN | 0.702025 | 3 | 0 | 3 |
| BAK1 | SLC29A1 | 0.701624 | 3 | 0 | 3 |
| BAK1 | FAM111B | 0.699949 | 3 | 0 | 3 |
For details and further investigation, click here